Abstract
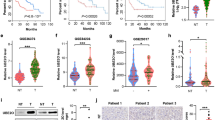
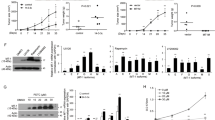
Methionine adenosyltransferase 1A (MAT1A) is a tumor suppressor downregulated in hepatocellular carcinoma and cholangiocarcinoma, two of the fastest rising cancers worldwide. We compared MATα1 (protein encoded by MAT1A) interactome in normal versus cancerous livers by mass spectrometry to reveal interactions with 14-3-3ζ. The MATα1/14-3-3ζ complex was critical for the expression of 14-3-3ζ. Similarly, the knockdown and small molecule inhibitor for 14-3-3ζ (BV02), and ChIP analysis demonstrated the role of 14-3-3ζ in suppressing MAT1A expression. Interaction between MATα1 and 14-3-3ζ occurs directly and is enhanced by AKT2 phosphorylation of MATα1. Blocking their interaction enabled nuclear MATα1 translocation and inhibited tumorigenesis. In contrast, overexpressing 14-3-3ζ lowered nuclear MATα1 levels and promoted tumor progression. However, tumor-promoting effects of 14-3-3ζ were eliminated when liver cancer cells expressed mutant MATα1 unable to interact with 14-3-3ζ. Taken together, the reciprocal negative regulation that MATα1 and 14-3-3ζ exert is a key mechanism in liver tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
19 August 2021
The name of Lucia Barbier- Torres was given incorrect in HTML version of the article. Correct is: Given name: Lucia Family name: Barbier-Torres
References
Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiological Rev. 2012;92:1515–42.
Yang H, Liu T, Wang J, Li TW, Fan W, Peng H, et al. Deregulated methionine adenosyltransferase α1, c-Myc, and Maf proteins together promote cholangiocarcinoma growth in mice and humans. Hepatology 2016;64:439–55.
Li Y, Lu L, Tu J, Zhang J, Xiong T, Fan W, et al. Reciprocal regulation between forkhead box M1/NF-kB and methionine adenosyltransferase 1A drives liver cancer. Hepatology 2020;72:1682–1700.
Torres L, Avila MA, Carretero MV, Latasa MU, Caballeria J, López-Rodas G, et al. Liver-specific methionine adenosyltransferase MAT1A gene expression is associated with a specific pattern of promoter methylation and histone acetylation: implications for MAT1A silencing during transformation. FASEB J. 2000;14:95–102.
Tomasi ML, Li TW, Li M, Mato JM, Lu SC. Inhibition of human methionine adenosyltransferase 1A transcription by coding region methylation. J Cell Physiol. 2012;227:1583–91.
Vázquez–Chantada M, Fernández–Ramos D, Embade N, Martínez–Lopez N, Varela–Rey M, Woodhoo A, et al. HuR/Methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation, and carcinogenesis. Gastroenterology 2010;138:1943–53.
Liu T, Yang H, Fan W, Tu J, Li TWH, Wang J, et al. Mechanisms of MAFG dysregulation in cholestatic liver injury and development of liver cancer. Gastroenterology 2018;155:557–71.
Murray B, Barbier-Torres L, Fan W, Mato JM, Lu SC. Methionine adenosyltransferases in liver cancer. World J Gastro. 2019;25:4300–19.
Yang H, Cho ME, Li TWH, Peng H, Ko KS, Mato JM, et al. MicroRNAs regulate methionine adenosyltransferase 1A expression in hepatocellular carcinoma. J Clin Invest. 2013;123:285–98.
Fan W, Yang H, Liu T, Wang J, Li TWH, Mavila N, et al. Prohibitin 1 suppresses liver cancer tumorigenesis in mice and human hepatocellular and cholangiocarcinoma cells. Hepatology 2017;65:1249–66.
Yang H, Li TWH, Peng J, Tang X, Ko KS, Xia M, et al. A mouse model of cholestasis-associated cholangiocarcinoma and transcription factors involved in progression. Gastroenterology 2011;141:378–88.
Xia Q, Li Z, Zheng J, Zhang X, Di Y, Ding J, et al. Identification of novel biomarkers for hepatocellular carcinoma using transcriptome analysis. J Cell Physiol. 2019;234:4851–63.
Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J. 2010;427:69–78.
Root A, Beizaei A, Ebhardt HA. Structure-based assessment and network analysis of targeting 14-3-3 proteins in prostate cancer. Mol Cancer. 2018;17:156.
Reytor E, Pérez-Miguelsanz J, Alvarez L, Pérez-Sala D, Pajares MA. Conformational signals in the C-terminal domain of methionine adenosyltransferase I/III determine its nucleocytoplasmic distribution. FASEB J. 2009;23:3347–60.
Pérez C, Pérez-Zúñiga FJ, Garrido F, Reytor E, Portillo F, Pajares MA. The oncogene PDRG1 is an interaction target of methionine adenosyltransferases. PLoS One. 2016;11:e0161672.
Murray B, Peng H, Barbier-Torres L, Robinson AE, Li TWH, Fan W, et al. Methionine adenosyltransferase α1 is targeted to the mitochondrial matrix and interacts with cytochrome P450 2E1 to lower its expression. Hepatology 2019;70:2018–34.
Frau M, Feo F, Pascale RM. Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis. J Hepatol. 2013;59:830–41.
Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 1996;84:889–97.
Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 1997;91:961–71.
Graves PR, Lovly CM, Uy GL, Piwnica-Worms H. Localization of human Cdc25C is regulated both by nuclear export and 14-3-3 protein binding. Oncogene 2001;20:1839–51.
Tang Y, Lv P, Sun Z, Han L, Luo B, Zhou W. 14-3-3ζ up-regulates hypoxia-inducible factor-1α in hepatocellular carcinoma via activation of PI3K/Akt/NF-кB signal transduction pathway. Int J Clin Exp Path. 2015;8:15845–53.
Ridder DA, Schindeldecker M, Weimann A, Berndt K, Urbansky L, Witzel HR, et al. Key enzymes in pyrimidine synthesis, CAD and CPS1, predict prognosis in hepatocellular carcinoma. Cancers 2021;13:744.
Liu H, Dong H, Robertson K, Liu C. DNA methylation suppresses expression of the urea cycle enzyme carbamoyl phosphate synthetase 1 (cps1) in human hepatocellular carcinoma. Am J Pathol. 2011;178:652–61.
Xu X, Sakon M, Nagano H, Hiraoka N, Yamamoto H, Hayashi N, et al. Akt2 expression correlates with prognosis of human hepatocellular carcinoma. Oncol Rep. 2004;11:25–32.
Wang C, Che L, Hu J, Zhang S, Jiang L, Latte G, et al. Activated mutant forms of PIK3CA cooperate with RasV12 or c-Met to induce liver tumour formation in mice via AKT2/mTORC1 cascade. Liver Int. 2016;36:1176–86.
Mroweh M, Roth G, Decaens T, Marche PN, Lerat H, Jílková ZM. Targeting Akt in hepatocellular carcinoma and its tumor microenvironment. Int J Mol Sci 2021;22:1794.
Ewald F, Grabinski N, Grottke A, Windhorst S, Nörz D, Carstensen L, et al. Combined targeting of AKT and mTOR using MK-2206 and RAD001 is synergistic in the treatment of cholangiocarcinoma. Int J Cancer 2013;133:2065–76.
Yu M, Guo HX, Hui C, Wang XH, Li CY, Zhan YQ, et al. 14-3-3ζ interacts with hepatocyte nuclear factor 1α and enhances its DNA binding and transcriptional activation. Biochimica et Biophysica Acta. 2013;1829:970–79.
Bergamaschi A, Christensen BL, Katzenellenbogen BS. Reversal of endocrine resistance in breast cancer: interrelationships among 14-3-3ζ, FOXM1, and a gene signature associated with mitosis. Breast Cancer Res. 2011;13:R70.
Su R, Gong JN, Chen MT, Song L, Shen C, Zhang XH, et al. c-Myc suppresses miR-451⊣YWTAZ/AKT axis via recruiting HDAC3 in acute myeloid leukemia. Oncotarget 2016;7:77430–43.
Peng H, Dara L, Li TWH, Zheng Y, Yang HP, Tomasi ML, et al. MAT2B-GIT1 interplay activates MEK1/ERK 1 and 2 to induce growth in human liver and colon cancer. Hepatology 2013;57:2299–313.
Acknowledgements
This work was supported by NIH grants DK123763 (HP Yang, JM Mato and SC Lu), P01CA233452 (HP Yang, E Seki, ML Tomasi, N Bhowmick, and SC Lu), Natural Science Foundation General Program of Hunan Province NO.2018JJ2664 (T Liu), and Plan Nacional of I+D SAF2017-88041-R (JM Mato). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
LL and JZ, data collection, analysis and interpretation, figure preparation and drafting of the manuscript; WF and YL, data collection, analysis, interpretation and figure preparation; JW, TWHL and LBT, cell culture, analysis and interpretation; JMM, ES, and NAB, critical reading, editing of manuscript and intellectual content. TL, provided human biospecimens; MM and MLT, technical assistance; HY, data collection, analysis, interpretation, figure preparation and drafting of the manuscript. SCL, study concept and design, data interpretation, edited the manuscript, obtained funding and provided overall study supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lu, L., Zhang, J., Fan, W. et al. Deregulated 14-3-3ζ and methionine adenosyltransferase α1 interplay promotes liver cancer tumorigenesis in mice and humans. Oncogene 40, 5866–5879 (2021). https://doi.org/10.1038/s41388-021-01980-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01980-6