Abstract
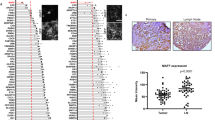
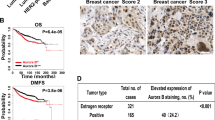
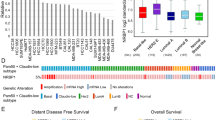
Metastatic breast cancer causes most breast cancer-associated deaths, especially in triple negative breast cancers (TNBC). The metastatic drivers of TNBCs are still poorly understood, and effective treatment non-existent. Here we reveal that the presence of Aurora-A Kinase (AURKA) in the nucleus and metastatic dissemination are molecularly connected through HIF1 (Hypoxia-Inducible Factor-1) signaling. Nuclear AURKA activates transcription of “hypoxia-induced genes” under normoxic conditions (pseudohypoxia) and without upregulation of oxygen-sensitive HIF1A subunit. We uncover that AURKA preferentially binds to HIF1B and co-localizes with the HIF complex on DNA. The mass-spectrometry analysis of the AURKA complex further confirmed the presence of CBP and p300 along with other TFIIB/RNApol II components. Importantly, the expression of multiple HIF-dependent genes induced by nuclear AURKA (N-AURKA), including migration/invasion, survival/death, and stemness, promote early cancer dissemination. These results indicate that nuclear, but not cytoplasmic, AURKA is a novel driver of early metastasis. Analysis of clinical tumor specimens revealed a correlation between N-AURKA presence and decreased patient survival. Our results establish a mechanistic link between two critical pathways in cancer metastasis, identifying nuclear AURKA as a crucial upstream regulator of the HIF1 transcription complex and a target for anti-metastatic therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Schumacher JM, Ashcroft N, Donovan PJ, Golden A. A highly conserved centrosomal kinase, AIR-1, is required for accurate cell cycle progression and segregation of developmental factors in Caenorhabditis elegans embryos. Development. 1998;125:4391–402.
Giet R, McLean D, Descamps S, Lee MJ, Raff JW, Prigent C, et al. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J Cell Biol. 2002;156:437–51.
Wang LH, Xiang J, Yan M, Zhang Y, Zhao Y, Yue CF, et al. The mitotic kinase Aurora-A induces mammary cell migration and breast cancer metastasis by activating the Cofilin-F-actin pathway. Cancer Res. 2010;70:9118–28.
Baba Y, Nosho K, Shima K, Irahara N, Kure S, Toyoda S, et al. Aurora-A expression is independently associated with chromosomal instability in colorectal cancer. Neoplasia. 2009;11:418–25.
Xu HT, Ma L, Qi FJ, Liu Y, Yu JH, Dai SD, et al. Expression of serine threonine kinase 15 is associated with poor differentiation in lung squamous cell carcinoma and adenocarcinoma. Pathol Int. 2006;56:375–80.
Zheng F, Yue C, Li G, He B, Cheng W, Wang X, et al. Nuclear AURKA acquires kinase-independent transactivating function to enhance breast cancer stem cell phenotype. Nat Commun. 2016;7:10180.
Tatsuka M, Sato S, Kanda A, Miki T, Kamata N, Kitajima S, et al. Oncogenic role of nuclear accumulated Aurora-A. Mol Carcinog. 2009;48:810–20.
Mao JH, Wu D, Perez-Losada J, Jiang T, Li Q, Neve RM, et al. Crosstalk between Aurora-A and p53: frequent deletion or downregulation of Aurora-A in tumors from p53 null mice. Cancer Cell. 2007;11:161–73.
Yang H, He L, Kruk P, Nicosia SV, Cheng JQ. Aurora-A induces cell survival and chemoresistance by activation of Akt through a p53-dependent manner in ovarian cancer cells. Int J Cancer. 2006;119:2304–12.
Briassouli P, Chan F, Savage K, Reis-Filho JS, Linardopoulos S. Aurora-A regulation of nuclear factor-kappaB signaling by phosphorylation of IkappaBalpha. Cancer Res. 2007;67:1689–95.
Xu J, Wu X, Zhou WH, Liu AW, Wu JB, Deng JY, et al. Aurora-A identifies early recurrence and poor prognosis and promises a potential therapeutic target in triple negative breast cancer. PLoS One. 2013;8:e56919.
D’Assoro AB, Liu T, Quatraro C, Amato A, Opyrchal M, Leontovich A, et al. The mitotic kinase Aurora-a promotes distant metastases by inducing epithelial-to-mesenchymal transition in ERalpha(+) breast cancer cells. Oncogene. 2014;33:599–610.
Nadler Y, Camp RL, Schwartz C, Rimm DL, Kluger HM, Kluger Y. Expression of Aurora A (but not Aurora B) is predictive of survival in breast cancer. Clin Cancer Res. 2008;14:4455–62.
Jiang BH, Rue E, Wang GL, Roe R, Semenza GL. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J Biol Chem. 1996;271:17771–8.
Semenza GL. The hypoxic tumor microenvironment: a driving force for breast cancer progression. Biochim Biophys Acta. 2016;1863:382–91.
De Bock K, Mazzone M, Carmeliet P. Antiangiogenic therapy, hypoxia, and metastasis: risky liaisons, or not? Nat Rev Clin Oncol. 2011;8:393–404.
Zhang H, Wong CC, Wei H, Gilkes DM, Korangath P, Chaturvedi P, et al. HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene. 2012;31:1757–70.
Hayashi Y, Yokota A, Harada H, Huang G. Hypoxia/pseudohypoxia-mediated activation of hypoxia-inducible factor-1alpha in cancer. Cancer Sci. 2019;110:1510–7.
Kozyreva VK, Kiseleva AA, Ice RJ, Jones BC, Loskutov YV, Matalkah F, et al. Combination of Eribulin and Aurora A inhibitor MLN8237 prevents metastatic colonization and induces cytotoxic autophagy in breast cancer. Mol Cancer Ther. 2016;15:1809–22.
Bierie B, Pierce SE, Kroeger C, Stover DG, Pattabiraman DR, Thiru P, et al. Integrin-beta4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc Natl Acad Sci USA. 2017;114:E2337–46.
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–8.
Yang N, Wang C, Wang Z, Zona S, Lin SX, Wang X, et al. FOXM1 recruits nuclear Aurora kinase A to participate in a positive feedback loop essential for the self-renewal of breast cancer stem cells. Oncogene 2017;36:3428–40.
Azab AK, Hu J, Quang P, Azab F, Pitsillides C, Awwad R, et al. Hypoxia promotes dissemination of multiple myeloma through acquisition of epithelial to mesenchymal transition-like features. Blood 2012;119:5782–94.
Melchionna R, Romani M, Ambrosino V, D’Arcangelo D, Cencioni C, Porcelli D, et al. Role of HIF-1alpha in proton-mediated CXCR4 down-regulation in endothelial cells. Cardiovasc Res. 2010;86:293–301.
Orlando IMC, Lafleur VN, Storti F, Spielmann P, Crowther L, Santambrogio S, et al. Distal and proximal hypoxia response elements cooperate to regulate organ-specific erythropoietin gene expression. Haematologica. 2020;105:2774–84.
Brooks DL, Schwab LP, Krutilina R, Parke DN, Sethuraman A, Hoogewijs D, et al. ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models. Mol Cancer. 2016;15:26.
Tanaka E, Hashimoto Y, Ito T, Okumura T, Kan T, Watanabe G, et al. The clinical significance of Aurora-A/STK15/BTAK expression in human esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11:1827–34.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49.
Andrzejewski S, Klimcakova E, Johnson RM, Tabaries S, Annis MG, McGuirk S, et al. PGC-1alpha promotes breast cancer metastasis and confers bioenergetic flexibility against metabolic drugs. Cell Metab. 2017;26:778–87 e5.
Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. 2011;23:55–64.
Pankova K, Rosel D, Novotny M, Brabek J. The molecular mechanisms of transition between mesenchymal and amoeboid invasiveness in tumor cells. Cell Mol Life Sci. 2010;67:63–71.
Rankin EB, Giaccia AJ. Hypoxic control of metastasis. Science 2016;352:175–80.
Chen A, Sceneay J, Godde N, Kinwel T, Ham S, Thompson EW, et al. Intermittent hypoxia induces a metastatic phenotype in breast cancer. Oncogene. 2018;37:4214–25.
Bos R, van der Groep P, Greijer AE, Shvarts A, Meijer S, Pinedo HM, et al. Levels of hypoxia-inducible factor-1alpha independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97:1573–81.
Gruber G, Greiner RH, Hlushchuk R, Aebersold DM, Altermatt HJ, Berclaz G, et al. Hypoxia-inducible factor 1 alpha in high-risk breast cancer: an independent prognostic parameter? Breast Cancer Res. 2004;6:R191–8.
Generali D, Berruti A, Brizzi MP, Campo L, Bonardi S, Wigfield S, et al. Hypoxia-inducible factor-1alpha expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer. Clin Cancer Res. 2006;12:4562–8.
Wang W, He YF, Sun QK, Wang Y, Han XH, Peng DF, et al. Hypoxia-inducible factor 1alpha in breast cancer prognosis. Clin Chim Acta. 2014;428:32–7.
Dupuy F, Tabaries S, Andrzejewski S, Dong Z, Blagih J, Annis MG, et al. PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab. 2015;22:577–89.
Pugacheva EN, Golemis EA. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol. 2005;7:937–46.
Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–63.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–31.
Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227.
Jones BC, Kelley LC, Loskutov YV, Marinak KM, Kozyreva VK, Smolkin MB, et al. Dual targeting of mesenchymal and amoeboid motility hinders metastatic behavior. Mol Cancer Res. 2017;15:670–82.
Ice RJ, McLaughlin SL, Livengood RH, Culp MV, Eddy ER, Ivanov AV, et al. NEDD9 depletion destabilizes Aurora A kinase and heightens the efficacy of Aurora A inhibitors: implications for treatment of metastatic solid tumors. Cancer Res. 2013;73:3168–80.
Acknowledgements
We thank WVU HSC Office of Graduate Research and Education for outstanding administrative support, WVU Tissue Bank, Genetic Tumor Modeling Core, and Animal Models & Imaging Facilities, supported by the WVUCI and NIH grants P20 RR016440, P30 RR032138/GM103488, and S10RR026378. Flow Cytometry Facility was supported by NIH grants P30GM103488, P30RR032138, and RCP1101809. Bioinformatics Core by NIH grants P20GM103434 and U54 GM-104942. This work was supported by a grant from NIH-NCI CA148671 to ENP. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number CA148671 (to ENP). Small animal imaging and cell image analysis were performed in the West Virginia University Animal Models & Imaging Facility, supported by NIH grants P20RR016440, P30GM103488, U54GM104942, P20GM103434, and S10RR026378. The flow cytometry analysis was done in WVU Flow Cytometry & Single Cell Core Facility, supported by NIH grants GM121322, GM104942, GM103434, and OD016165. The genetic tumor modeling (GTM) core facility provided animal models supported by NIH grant GM121322. The WVU Bioinformatics Core Facility supported by NIH grants P20GM103434 and U54 GM-104942 assisted with RNA-Seq data analysis and interpretation.
Author information
Authors and Affiliations
Contributions
KMW and ENP conceived the project and wrote the manuscript. TMA statistical analysis was performed by ENP. Animal pathology and tumor analysis was done by MSS and KMW. Bioinformatics analysis was performed by DC, GH, JG, and KMW. All remaining experiments and data analysis were performed by KMW, AM, MAV, JL, HC, SY, NVM, SW, MLP and ENP.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Whately, K.M., Voronkova, M.A., Maskey, A. et al. Nuclear Aurora-A kinase-induced hypoxia signaling drives early dissemination and metastasis in breast cancer: implications for detection of metastatic tumors. Oncogene 40, 5651–5664 (2021). https://doi.org/10.1038/s41388-021-01969-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01969-1
This article is cited by
-
ROS production by mitochondria: function or dysfunction?
Oncogene (2024)
-
Exploring Prognostic Gene Factors in Breast Cancer via Machine Learning
Biochemical Genetics (2024)
-
Controlled tumor heterogeneity in a co-culture system by 3D bio-printed tumor-on-chip model
Scientific Reports (2023)