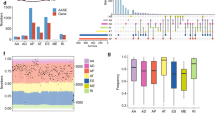
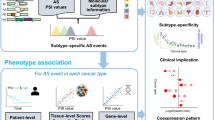
Abstract
Aberrant alternative splicing events (AASEs) are key biological processes for tumorigenesis and the rationale for designing splice-switching oligonucleotides (SSOs). However, the landscape of AASEs in esophageal squamous cell carcinoma (ESCC) remains unclear, which undermines the development of SSOs for ESCC. Here, we profiled AASEs based on 125 pairs of RNA-seq libraries. We identified 14,710 AASEs in ESCC, most of which (92.67%) affected coding genes. The first exon of transcripts was frequently changed in ESCC. We constructed a regulatory network where 74 RNA-binding proteins regulated 2142 AASEs. This network was enriched in apoptotic pathways and various adhesion/junction-related processes. Somatic mutations in ESCC regulating ASEs were mainly through trans-regulatory mode and were enriched in intron regions. Isoform switches of apoptotic genes and binding genes both tended to induce “noncoding transcripts” and “domain loss,” disrupting the apoptotic and Hippo signaling pathways. All ESCC samples were grouped into three clusters with different AASEs patterns and the second cluster was identified as “cold tumor,” with a low abundance of immune cells, activated immune pathways, and immunomodulators. Our work comprehensively profiled the landscape of AASEs in ESCC, revealed novel AASEs related to tumorigenesis and immune microenvironment, and suggested promising directions for designing SSOs for ESCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Genomic and transcriptomic data are available under accession no. PRJCA002085 in the Genome Sequence Archive of Beijing Institute of Genomics, Chinese Academy of Sciences.
References
Mao S, Li Y, Lu Z, Che Y, Sun S, Huang J, et al. Survival-associated alternative splicing signatures in esophageal carcinoma. Carcinogenesis. 2019;40:121–30.
Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet. 2017;390:2383–96.
Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499–509.
Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–12.
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6.
Pan Q, Shai O, Lee LJ, Frey J, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5.
Chabot B, Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol. 2016;212:13–27.
Halloy F, Iyer PS, Ćwiek P, Ghidini A, Barman-Aksözen J, Wildner-Verhey van Wijk N, et al. Delivery of oligonucleotides to bone marrow to modulate ferrochelatase splicing in a mouse model of erythropoietic protoporphyria. Nucleic Acids Res. 2020;48:4658–71.
O K, C W. Oligonucleotide therapies for disorders of the nervous system. Nat Biotechnol. 2017;35:249–63.
Michaels WE, Bridges RJ, Hastings ML. Antisense oligonucleotide-mediated correction of CFTR splicing improves chloride secretion in cystic fibrosis patient-derived bronchial epithelial cells. Nucleic Acids Res. 2020;48:7454–67.
Wang L, Kempton JB, Jiang H, Jodelka FM, Brigande AM, Dumont RA, et al. Fetal antisense oligonucleotide therapy for congenital deafness and vestibular dysfunction. Nucleic Acids Res. 2020;48:5065–80.
Mogilevsky M, Shimshon O, Kumar S, Mogilevsky A, Keshet E, Yavin E, et al. Modulation of MKNK2 alternative splicing by splice-switching oligonucleotides as a novel approach for glioblastoma treatment. Nucleic Acids Res. 2018;46:11396–404.
Li L, Hobson L, Perry L, Clark B, Heavey S, Haider A, et al. Targeting the ERG oncogene with splice-switching oligonucleotides as a novel therapeutic strategy in prostate cancer. Br J Cancer. 2020;123:1024–32.
Brandt AC, McNally L, Lorimer EL, Unger B, Koehn OJ, Suazo KF, et al. Splice switching an oncogenic ratio of SmgGDS isoforms as a strategy to diminish malignancy. Proc Natl Acad Sci USA. 2020;117:3627–36.
ST C, JL W, CF B, BF B. RNA-targeted therapeutics. Cell Metab. 2018;27:714–39.
J D, P B-G, M C-F. Targeting mRNA processing as an anticancer strategy. Nat Rev Drug Discov. 2020;19:112–29.
Matlin AJ, Clark F, Smith CWJ. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–98.
Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18.
Bonnal SC, López-Oreja I, Valcárcel J. Roles and mechanisms of alternative splicing in cancer - implications for care. Nat Rev Clinical oncology. 2020;17:457–74.
Garrido-Martín D, Borsari B, Calvo M, Reverter F, Guigó R. Identification and analysis of splicing quantitative trait loci across multiple tissues in the human genome. Nat Commun. 2021;12:727.
Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:660.
Boise LH, González-García M, Postema CE, Ding L, Lindsten T, Turka LA, et al. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608.
Frankiw L, Baltimore D, Li G. Alternative mRNA splicing in cancer immunotherapy. Nat Rev Immunol. 2019;19:675–87.
Xie ZC, Wu HY, Ma FC, Dang YW, Peng ZG, Zhou HF, et al. Prognostic alternative splicing signatures and underlying regulatory network in esophageal carcinoma. Am J Transl Res. 2019;11:4010–28.
Li SL, Hu ZX, Zhao YJ, Huang SL, He XH. Transcriptome-wide analysis reveals the landscape of aberrant alternative splicing events in liver cancer. Hepatology. 2019;69:359–75.
Vanharanta S, Marney CB, Shu W, Valiente M, Zou Y, Mele A, et al. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. eLife. 2014;3:e02734
Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu HZ, Shen SH, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29:3286–300.
Akgul C, Moulding DA, Edwards SW. Alternative splicing of Bcl-2-related genes: functional consequences and potential therapeutic applications. Cell Mol Life Sci. 2004;61:2189–99.
Peterson RE, Hoffman S, Kern MJ. Opposing roles of two isoforms of the Prx1 homeobox gene in chondrogenesis. Dev Dyn. 2005;233:811–21.
Soliman MA, Berardi P, Pastyryeva S, Bonnefin P, Feng X, Colina A, et al. ING1a expression increases during replicative senescence and induces a senescent phenotype. Aging Cell. 2008;7:783–94.
Liu Y, Huang W, Gao X, Kuang F. Regulation between two alternative splicing isoforms ZNF148(FL) and ZNF148(ΔN), and their roles in the apoptosis and invasion of colorectal cancer. Pathol, Res Pract. 2019;215:272–7.
Lucena-Araujo AR, Kim HT, Thome C, Jacomo RH, Melo RA, Bittencourt R, et al. High delta Np73/TAp73 ratio is associated with poor prognosis in acute promyelocytic leukemia. Blood. 2015;126:2302–6.
Aigner P, Mizutani T, Horvath J, Eder T, Heber S, Lind K, et al. STAT3β is a tumor suppressor in acute myeloid leukemia. Blood Adv. 2019;3:1989–2002.
Liu M, An H, Zhang Y, Sun W, Cheng S, Wang R, et al. Molecular analysis of Chinese oesophageal squamous cell carcinoma identifies novel subtypes associated with distinct clinical outcomes. EBioMedicine. 2020;57:102831.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60.
Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 2012;22:1760–74.
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290.
Trincado JL, Entizne JC, Hysenaj G, Singh B, Skalic M, Elliott DJ, et al. SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018;19:40
Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33:831.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Ryan M, Wong WC, Brown R, Akbani R, Su X, Broom B, et al. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 2016;44:D1018–22.
Ryan MC, Cleland J, Kim R, Wong WC, Weinstein JN. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics. 2012;28:2385–7.
Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–8.
Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Vitting-Seerup K, Sandelin A. IsoformSwitchAnalyzeR: analysis of changes in genome-wide patterns of alternative splicing and its functional consequences. Bioinformatics. 2019;35:4469–71.
Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2011;40:D290–301.
Kang Y-J, Yang D-C, Kong L, Hou M, Meng Y-Q, Wei L, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45:W12–6.
Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–3.
Klausen MS, Jespersen MC, Nielsen H, Jensen KK, Jurtz VI, Sønderby CK, et al. NetSurfP-2.0: improved prediction of protein structural features by integrated deep learning. 2018. https://doi.org/10.1101/311209.
de la Fuente L, Arzalluz-Luque Á, Tardáguila M, Tardáguila M, del Risco H, Martí C, et al. tappAS: a comprehensive computational framework for the analysis of the functional impact of differential splicing. 2019. https://doi.org/10.1101/690743.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7.
Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35:i436–45.
Finotello F, Mayer C, Plattner C, Laschober G, Rieder D, Hackl H, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11:34.
Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife. 2017;6:e26476.
Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220.
Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174.
Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218.
Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013;14:7.
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The immune landscape of cancer. Immunity. 2018;48:812–30.e814.
Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3.
Acknowledgements
This research was supported by the Research Fund of Key Laboratory of Xinjiang oncology (Grant no. 2017D04006). Funders only provided funding and had no role in the study design, data collection, data analysis, interpretation, and writing of the report. The authors thank Dr Wen Zhang (Department of Immunology, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China) for providing technical assistance.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, Q., Zhang, Y., An, H. et al. The landscape and biological relevance of aberrant alternative splicing events in esophageal squamous cell carcinoma. Oncogene 40, 4184–4197 (2021). https://doi.org/10.1038/s41388-021-01849-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01849-8
This article is cited by
-
Multi-omics analysis reveals RNA splicing alterations and their biological and clinical implications in lung adenocarcinoma
Signal Transduction and Targeted Therapy (2022)