Abstract
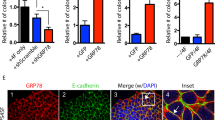
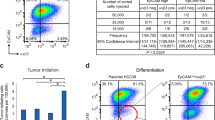
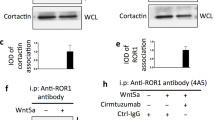
The heat shock protein GRP78 typically resides in the endoplasmic reticulum in normal tissues, but it has been shown to be expressed on the cell surface of several cancer cells, and some stem cells, where it can act as a signaling molecule by not-yet-fully defined mechanisms. Although cell surface GRP78 (sGRP78) has emerged as an attractive chemotherapeutic target, understanding how sGRP78 is functioning in cancer has been complicated by the fact that sGRP78 can function in a cell-context dependent manner, with a diverse array of reported binding partners, to regulate a variety of cellular responses. We had previously shown that sGRP78 was important in regulating pluripotent stem cell (PSC) functions, and hypothesized that embryonic-like mechanisms of GRP78 were critical to regulating aggressive breast cancer cell functions. Here, using proteomics we identify Dermcidin (DCD) as a novel sGRP78 binding partner common to both PSCs and breast cancer cells. We show that GRP78 and DCD cooperate to regulate stem cell and cancer cell migration that is dependent on the cell surface functions of these proteins. Finally, we identify Wnt/β-catenin signaling, a critical pathway in stem cell and cancer cell biology, as an important downstream intermediate in regulating this migration phenotype.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507.
Holm F, Hellqvist E, Mason CN, Ali SA, Delos-Santos N, Barrett CL, et al. Reversion to an embryonic alternative splicing program enhances leukemia stem cell self-renewal. Proc Natl Acad Sci U.S.A. 2015;112:15444–9.
Mizuno H, Spike BT, Wahl GM, Levine AJ. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc Natl Acad Sci U.S.A. 2010;107:22745–50.
Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN. et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173:338–54.
Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67:3496–9.
Lee AS. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat Rev Cancer. 2014;14:263–6.
Conner C, Lager TW, Guldner IH, Wu MZ, Hishida Y, Hishida T, et al. Cell surface GRP78 promotes stemness in normal and neoplastic cells. Sci Rep. 2020;10:3474.
Kelber JA, Panopoulos AD, Shani G, Booker EC, Belmonte JC, Vale WW, et al. Blockade of Cripto binding to cell surface GRP78 inhibits oncogenic Cripto signaling via MAPK/PI3K and Smad2/3 pathways. Oncogene. 2009;28:2324–36.
Miharada K, Karlsson G, Rehn M, Rorby E, Siva K, Cammenga J, et al. Cripto regulates hematopoietic stem cells as a hypoxic-niche-related factor through cell surface receptor GRP78. Cell Stem Cell. 2011;9:330–44.
Spike BT, Kelber JA, Booker E, Kalathur M, Rodewald R, Lipianskaya J, et al. CRIPTO/GRP78 signaling maintains fetal and adult mammary stem cells ex vivo. Stem Cell Rep. 2014;2:427–39.
Tsai YL, Zhang Y, Tseng CC, Stanciauskas R, Pinaud F, Lee AS. Characterization and mechanism of stress-induced translocation of 78-kilodalton glucose-regulated protein (GRP78) to the cell surface. J Biol Chem. 2015;290:8049–64.
Davidson DJ, Haskell C, Majest S, Kherzai A, Egan DA, Walter KA, et al. Kringle 5 of human plasminogen induces apoptosis of endothelial and tumor cells through surface-expressed glucose-regulated protein 78. Cancer Res. 2005;65:4663–72.
Tsai YL, Ha DP, Zhao H, Carlos AJ, Wei S, Pun TK, et al. Endoplasmic reticulum stress activates SRC, relocating chaperones to the cell surface where GRP78/CD109 blocks TGF-beta signaling. Proc Natl Acad Sci U.S.A. 2018;115:E4245–54.
Tseng CC, Stanciauskas R, Zhang P, Woo D, Wu K, Kelly K, et al. GRP78 regulates CD44v membrane homeostasis and cell spreading in tamoxifen-resistant breast cancer. Life Sci Alliance. 2019;2:e201900377.
Schittek B. The multiple facets of dermcidin in cell survival and host defense. J Innate Immun. 2012;4:349–60.
Porter D, Weremowicz S, Chin K, Seth P, Keshaviah A, Lahti-Domenici J, et al. A neural survival factor is a candidate oncogene in breast cancer. Proc Natl Acad Sci U.S.A. 2003;100:10931–6.
Stewart GD, Skipworth RJ, Ross JA, Fearon K, Baracos VE. The dermcidin gene in cancer: role in cachexia, carcinogenesis and tumour cell survival. Curr Opin Clin Nutr Metab Care. 2008;11:208–13.
Bancovik J, Moreira DF, Carrasco D, Yao J, Porter D, Moura R, et al. Dermcidin exerts its oncogenic effects in breast cancer via modulation of ERBB signaling. BMC Cancer. 2015;15:70.
Dunn KW, Kamocha MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300:C723–42.
Manders EMM, Verbeek FJ, Aten JA. Measurement of co‐localization of objects in dual‐colour confocal images. J Microsc. 1993;169:375–82.
Subik K, Lee JF, Baxter L, Strzepek T, Costello D, Crowley P, et al. The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast Cancer (Auckl). 2010;4:35–41.
Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–73.
Matsuda Y, Schlange T, Oakeley EJ, Boulay A, Hynes NE. WNT signaling enhances breast cancer cell motility and blockade of the WNT pathway by sFRP1 suppresses MDA-MB-231 xenograft growth. Breast Cancer Res. 2009;11:R32.
Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–7.
Liu GH, Barkho BZ, Ruiz S, Diep D, Qu J, Yang SL, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221–5.
Panopoulos AD, Ruiz S, Yi F, Herrerias A, Batchelder EM, Izpisua, et al. Rapid and highly efficient generation of induced pluripotent stem cells from human umbilical vein endothelial cells. PLoS One. 2011;6:e19743.
Acknowledgements
This work was supported in part by an American Cancer Society Research Scholar Grant (RSG-20-022-01-CDD), a Walther Cancer Foundation Advancing Basic Cancer grant, and by grant UL1TR001108 from the Indiana Clinical and Translational Sciences Institute (to ADP). CC was supported in part by grant TL1TR001107 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award. CRK was supported in part through a Hiller Family Research Fellowship. We thank the members of the D’Souza-Schorey laboratory for kindly providing valuable advice for the TOPFlash assays; Drs. Michelle Joyce, Matthew Champion, and Bill Boggess of the Notre Dame Mass Spectrometry and Proteomics Facility for proteomics; and Dr. Sara Cole of the Optical Microscopy Core, Notre Dame Integrated Imaging Facility (NDIIF), for her microscopy assistance and expertise. We thank Drs. Jonathan Kelber and Peter Gray for valuable discussions, and three anonymous reviewers for their helpful comments. We are grateful to the Gallagher Family for their generous support of stem cell research at the University of Notre Dame.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lager, T.W., Conner, C., Keating, C.R. et al. Cell surface GRP78 and Dermcidin cooperate to regulate breast cancer cell migration through Wnt signaling. Oncogene 40, 4050–4059 (2021). https://doi.org/10.1038/s41388-021-01821-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01821-6
This article is cited by
-
Unveiling the dark side of glucose-regulated protein 78 (GRP78) in cancers and other human pathology: a systematic review
Molecular Medicine (2023)
-
Autophagy, molecular chaperones, and unfolded protein response as promoters of tumor recurrence
Cancer and Metastasis Reviews (2023)
-
The Wnt/β-catenin signalling pathway in Haematological Neoplasms
Biomarker Research (2022)
-
Differential enrichment of H3K9me3 in intrahepatic cholangiocarcinoma
BMC Medical Genomics (2022)