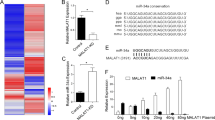
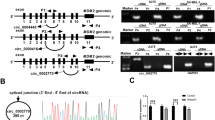
Abstract
MicroRNAs and RNA-binding proteins (RBPs) primarily target the 3′ UTR of mRNAs to control their translation and stability. However, their co-regulatory effects on specific mRNAs in physiology and disease are yet to be fully explored. CSDE1 is an RBP that promotes metastasis in melanoma and mechanisms underlying its oncogenic activities need to be completely defined. Here we report that CSDE1 interacts with specific miRNA-induced silencing complexes (miRISC) in melanoma. We find an association of CSDE1 with AGO2, the essential component of miRISC, which is facilitated by target mRNAs and depends on the first cold shock domain of CSDE1. Both CSDE1 and AGO2 bind to 3′ UTR of PMEPA1. CSDE1 counters AGO2 binding, leading to an increase of PMEPA1 expression. We also identify a miRNA, miR-129-5p, that represses PMEPA1 expression in melanoma. Collectively, our results show that PMEPA1 promotes tumorigenic traits and that CSDE1 along with miR-129-5p/AGO2 miRISC act antagonistically to fine-tune PMEPA1 expression toward the progression of melanoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105.
Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–15.
Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004.
Schadendorf D, Fisher DE, Garbe C, Gershenwald JE, Grob JJ, Halpern A, et al. Melanoma. Nat Rev Dis Prim. 2015;1:15003.
Xu Y, Brenn T, Brown ERS, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer. 2012;106:553–61.
Aksenenko M, Palkina N, Komina A, Tashireva L, Ruksha T. Differences in microRNA expression between melanoma and healthy adjacent skin. BMC Dermatol. 2019;19:1.
Qian H, Yang C, Yang Y. MicroRNA-26a inhibits the growth and invasiveness of malignant melanoma and directly targets on MITF gene. Cell Death Disco. 2017;3:17028.
Fattore L, Ruggiero CF, Pisanu ME, Liguoro D, Cerri A, Costantini S, et al. Reprogramming miRNAs global expression orchestrates development of drug resistance in BRAF mutated melanoma. Cell Death Differ. 2019;26:1267–82.
Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37.
Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:1–13.
Quévillon Huberdeau M, Simard MJ. A guide to microRNA-mediated gene silencing. FEBS J. 2019;286:642–52.
Nam JW, Rissland OS, Koppstein D, Abreu-Goodger C, Jan CH, Agarwal V, et al. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol Cell. 2014;53:1031–43.
van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–56.
Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer. 2017;3:506–28.
Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–43.
Kishore S, Luber S, Zavolan M. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief Funct Genomics. 2010;9:391–404.
Jiang P, Coller H. Functional interactions between micro-RNAs and RNA binding proteins. Microrna. 2012;1:70–79.
Iadevaia V, Gerber AP. Combinatorial control of mRNA fates by RNA-binding proteins and non-coding RNAs. Biomol Ther. 2015;5:2207–22.
Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–24.
Ahuja D, Goyal A, Ray PS. Interplay between RNA-binding protein HuR and microRNA-125b regulates p53 mRNA translation in response to genotoxic stress. RNA Biol. 2016;13:1152–65.
Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–8.
Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. Pumilio-induced RNA structure switch in p27-3’ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–20.
Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 2012;40:5088–100.
Li Y, Estep JA, Karginov FV. Transcriptome wide Identification and Validation of Interactions between the miRNA Machinery and HuR on mRNA Targets. J Mol Biol. 2017;430:285–96.
Sternburg EL, Estep JA, Nguyen DK, Li Y, Karginov FV. Antagonistic and co-operative AGO2-PUM interactions in regulating mRNAs. Sci Rep. 2018;8:15316.
Kelly TJ, Suzuki HI, Zamudio JR, Suzuki M, Sharp PA. Sequestration of microRNA-mediated target repression by the Ago2-associated RNA binding protein FAM120A. RNA. 2019;25:1291–7.
Mihailovich M, Militti C, Gabaldón T, Gebauer F. Eukaryotic cold shock domain proteins: highly versatile regulators of gene expression. BioEssays. 2010;32:109–18.
Hollmann NM, Jagtap PKA, Masiewicz P, Guitart T, Simon B, Provaznik J, et al. Pseudo-RNA-binding domains mediate RNA structure specificity in upstream of N-Ras. Cell Rep. 2020;32:107930.
Abaza I, Coll O, Patalano S, Gebauer F. Drosophila UNR is required for translational repression of male-specific lethal 2 mRNA during regulation of X-chromosome dosage compensation. Genes Dev. 2006;20:380–9.
Duncan K, Grskovic M, Strein C, Beckmann K, Niggeweg R, Abaza I, et al. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: translational repression for dosage compensation. Genes Dev. 2006;20:368–79.
Dormoy-Raclet V, Markovits J, Jacquemin-Sablon A, Jacquemin-Sablon H. Regulation of Unr expression by 5′- and 3′-untranslated regions of its mRNA through modulation of stability and IRES mediated translation. RNA Biol. 2005;2:e27–35.
Schepens B, Tinton SA, Bruynooghe Y, Parthoens E, Haegman M, Beyaert R, et al. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. EMBO J. 2007;26:158–69.
Mitchell SA, Brown EC, Coldwell MJ, Jackson RJ, Willis AE. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol Cell Biol. 2001;21:3364–74.
Cornelis S, Tinton SA, Schepens B, Bruynooghe Y, Beyaert R. UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein. Nucleic Acids Res. 2005;33:3095–108.
Dinur M, Kilav R, Sela-Brown A, Jacquemin-Sablon H, Naveh-Many T. In vitro evidence that upstream of N-ras participates in the regulation of parathyroid hormone messenger ribonucleic acid stability. Mol Endocrinol. 2006;20:1652–60.
Lee HJ, Bartsch D, Xiao C, Guerrero S, Ahuja G, Schindler C, et al. A post-transcriptional program coordinated by CSDE1 prevents intrinsic neural differentiation of human embryonic stem cells. Nat Commun. 2017;8:1456.
Elatmani H, Dormoy-Raclet V, Dubus P, Dautry F, Chazaud C, Jacquemin-Sablon H. The RNA-binding protein Unr prevents mouse embryonic stem cells differentiation toward the primitive endoderm lineage. Stem Cell Rep. 2011;29:1504–16.
Kakumani PK, Harvey LM, Houle F, Guitart T, Gebauer F, Simard MJ. CSDE1 controls gene expression through the miRNA-mediated decay machinery. Life Sci Alliance. 2020;3:e201900632.
Wurth L, Papasaikas P, Olmeda D, Bley N, Calvo GT, Guerrero S, et al. UNR/CSDE1 drives a post-transcriptional program to promote melanoma invasion and metastasis. Cancer Cell. 2016;30:694–707.
Fournier PG, Juárez P, Jiang G, Clines GA, Niewolna M, Kim HS, et al. The TGF-β signaling regulator PMEPA1 suppresses prostate cancer metastases to bone. Cancer Cell. 2015;27:809–21.
Amalia R, Abdelaziz M, Puteri MU, Hwang J, Anwar F, Watanabe Y, et al. TMEPAI/PMEPA1 inhibits Wnt signaling by regulating β-catenin stability and nuclear accumulation in triple negative breast cancer cells. Cell Signal. 2019;59:24–33.
Abdelaziz M, Watanabe Y, Kato M. PMEPA1/TMEPAI knockout impairs tumour growth and lung metastasis in MDA-MB-231 cells without changing monolayer culture cell growth. J Biochem. 2019;165:411–4.
Vo Nguyen TT, Watanabe Y, Shiba A, Noguchi M, Itoh S, Kato M. TMEPAI/PMEPA1 enhances tumorigenic activities in lung cancer cells. Cancer Sci. 2014;105:334–41.
Connerty P, Ahadi A, Hutvagner G. RNA binding proteins in the miRNA pathway. Int J Mol Sci. 2015;17:E31.
Jannot G, Vasquez-Rifo A, Simard MJ. Argonaute pull-down and RISC analysis using 2’-O-methylated oligonucleotides affinity matrices. Methods Mol Biol. 2011;725:233–49.
Golden RJ, Chen B, Li T, Braun J, Manjunath H, Chen X, et al. An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature. 2017;542:197–202.
Quévillon Huberdeau M, Zeitler DM, Hauptmann J, Bruckmann A, Fressigné L, Danner J, et al. Phosphorylation of Argonaute proteins affects mRNA binding and is essential for microRNA-guided gene silencing in vivo. EMBO J. 2017;36:2088–106.
Karginov FV, Hannon GJ. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013;27:1624–32.
Feng S, Zhu X, Fan B, Xie D, Li T, Zhang X. miR‑19a‑3p targets PMEPA1 and induces prostate cancer cell proliferation, migration and invasion. Mol Med Rep. 2016;13:4030–8.
Agarwal V, Bell GW, Nam J, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005.
Abaza I, Gebauer F. Functional domains of Drosophila UNR in translational control. RNA. 2008;14:482–90.
Kamenska A, Simpson C, Vindry C, Broomhead H, Bénard M, Ernoult-Lange M, et al. The DDX6-4E-T interaction mediates translational repression and P-body assembly. Nucleic Acids Res. 2016;44:6318–34.
Ji J, Ding K, Luo T, Xu R, Zhang X, Huang B, et al. PMEPA1 isoform a drives progression of glioblastoma by promoting protein degradation of the Hippo pathway kinase LATS1. Oncogene. 2020;39:1125–39.
Militti C, Maenner S, Becker PB, Gebauer F. UNR facilitates the interaction of MLE with the lncRNA roX2 during Drosophila dosage compensation. Nat Commun. 2014;5:4762.
Chang TC, Yamashita A, Chen CY, Yamashita Y, Zhu W, Durdan S, et al. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 2004;18:2010–23.
Kaye JA, Rose NC, Goldsworthy B, Goga A, L’Etoile ND. A 3′UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron. 2009;61:57–70.
Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat Struct Mol Biol. 2012;19:176–83.
Bohn JA, Van Etten JL, Schagat TL, Bowman BM, McEachin RC, Freddolino PL, et al. Identification of diverse target RNAs that are functionally regulated by human Pumilio proteins. Nucleic Acids Res. 2018;46:362–86.
Fishbein L, Leshchiner I, Walter V, Danilova L, Robertson AG, Johnson AR, et al. Comprehensive molecular characterization of pheochromocytoma and paraganglioma. Cancer Cell. 2017;31:181–93.
Martinez-Useros J, Garcia-Carbonero N, Li W, Fernandez-Aceñero MJ, Cristobal I, Rincon R, et al. UNR/ CSDE1 expression is critical to maintain invasive phenotype of colorectal cancer through regulation of c-MYC and epithelial-to-mesenchymal transition. J Clin Med. 2019;8:560.
Young LE, Moore AE, Sokol L, Meisner-Kober N, Dixon DA. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol Cancer Res. 2012;10:167–80.
Bottini S, Hamouda-Tekaya N, Mategot R, Zaragosi LE, Audebert S, Pisano S. et al. Post-transcriptional gene silencing mediated by microRNAs is controlled by nucleoplasmic Sfpq. Nat Commun. 2017;8:1189
Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96.
Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2017;131:1273–86.
Liu Q, Jiang J, Fu Y, Liu T, Yu Y, Zhang X. MiR-129-5p functions as a tumor suppressor in gastric cancer progression through targeting ADAM9. Biomed Pharmacother. 2018;105:420–7.
Wang S, Chen Y, Yu X, Lu Y, Wang H, Wu F, et al. miR-129-5p attenuates cell proliferation and epithelial mesenchymal transition via HMGB1 in gastric cancer. Pathol Res Pr. 2019;215:676–82.
Qiu Z, Wang X, Shi Y, Da M. miR-129-5p suppresses proliferation, migration, and induces apoptosis in pancreatic cancer cells by targeting PBX3. Acta Biochim Biophys Sin (Shanghai). 2019;51:997–1007.
Li Z, Lu J, Zeng G, Pang J, Zheng X, Feng J, et al. MiR-129-5p inhibits liver cancer growth by targeting calcium calmodulin-dependent protein kinase IV (CAMK4). Cell Death Dis. 2019;10:789.
Wan P, Bai X, Yang C, He T, Luo L, Wang Y, et al. miR-129-5p inhibits proliferation, migration, and invasion in rectal adenocarcinoma cells through targeting E2F7. J Cell Physiol. 2020;235:5689–701.
Li G, Xie J, Wang J. Tumor suppressor function of miR-129-5p in lung cancer. Oncol Lett. 2019;17:5777–83.
Ma L, Chen X, Li C, Cheng R, Gao Z, Meng X, et al. miR-129-5p and -3p co-target WWP1 to suppress gastric cancer proliferation and migration. J Cell Biochem. 2018;120:7527–38.
Goyer B, Pereira U, Magne B, Larouche D, Kearns‐Turcotte S, Rochette PJ, et al. Impact of ultraviolet radiation on dermal and epidermal DNA damage in a human pigmented bilayered skin substitute. J Tissue Eng Regen Med. 2019;13:2300–11.
Acknowledgements
We thank all members of our laboratory for helpful discussions and the Bioinformatics Unit at CRG Barcelona for their services in analyzing the CLiP-Seq data.
Funding
The Canadian Institutes of Health Research (CIHR) supported this work. PKK and LMH are recipients of scholarship from Fonds de Recherche du Québec-Santé (FRQ-S). MJS is a Research Chair from FRQ-S. LG was supported by the CIHR (FDN-143213). FG was supported by grants from the Spanish Ministry of Science and Innovation (MICINN, PGC2018-099697-B-I00), “la Caixa” Foundation (ID:100010434) under the agreement LCF/PR/HR17/52150016, the Catalan Government (2017SGR534) and the Center of Excellence Severo Ochoa.
Author information
Authors and Affiliations
Contributions
PKK designed and performed most of the experiments. TG and FH performed cancer assays, while LMH helped in the preparation of different reagents. TG also generated data for Fig. 3D, E. MJS and FG supervised the work. BG isolated, cultured, and prepared proteins from melanocytes used in Fig. 3E and was supervised by LG. PKK and MJS wrote the manuscript with the help of FG.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kakumani, P.K., Guitart, T., Houle, F. et al. CSDE1 attenuates microRNA-mediated silencing of PMEPA1 in melanoma. Oncogene 40, 3231–3244 (2021). https://doi.org/10.1038/s41388-021-01767-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01767-9