Abstract
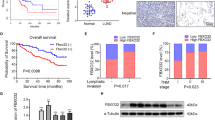
Mutations in the epidermal growth factor receptor (EGFR) tyrosine kinase domain constitutively activate EGFR resulting in lung tumorigenesis. Activated EGFR modulates downstream signaling by altering phosphorylation-driven interactions that promote growth and survival. Secretory carrier membrane proteins (SCAMPs) are a family of transmembrane proteins that regulate recycling of receptor proteins, including EGFR. The potential role of SCAMPs in mutant EGFR function and tumorigenesis has not been elucidated. Using quantitative mass-spectrometry-based phosphoproteomics, we identified SCAMP3 as a target of mutant EGFRs in lung adenocarcinoma and sought to further investigate the role of SCAMP3 in the regulation of lung tumorigenesis. Here we show that activated EGFR, either directly or indirectly phosphorylates SCAMP3 at Y86 and this phosphorylation increases the interaction of SCAMP3 with both wild-type and mutant EGFRs. SCAMP3 knockdown increases lung adenocarcinoma cell survival and increases xenograft tumor growth in vivo, demonstrating a tumor suppressor role of SCAMP3 in lung tumorigenesis. The tumor suppressor function is a result of SCAMP3 promoting EGFR degradation and attenuating MAP kinase signaling pathways. SCAMP3 knockdown also increases multinucleated cells in culture, suggesting that SCAMP3 is required for efficient cytokinesis. The enhanced growth, increased colony formation, reduced EGFR degradation and multinucleation phenotype of SCAMP3-depleted cells were reversed by re-expression of wild-type SCAMP3, but not SCAMP3 Y86F, suggesting that Y86 phosphorylation is critical for SCAMP3 function. Taken together, the results of this study demonstrate that SCAMP3 functions as a novel tumor suppressor in lung cancer by modulating EGFR signaling and cytokinesis that is partly Y86 phosphorylation-dependent.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000.
Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28:S24–31.
Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39.
Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500.
Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73.
Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–46.
Stewart EL, Tan SZ, Liu G, Tsao MS. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Transl Lung Cancer Res. 2015;4:67–81.
Maity TK, Venugopalan A, Linnoila I, Cultraro CM, Giannakou A, Nemati R, et al. Loss of MIG6 accelerates initiation and progression of mutant epidermal growth factor receptor-driven lung adenocarcinoma. Cancer Discov. 2015;5:534–49.
Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–84.
Zhang X, Belkina N, Jacob HK, Maity T, Biswas R, Venugopalan A, et al. Identifying novel targets of oncogenic EGF receptor signaling in lung cancer through global phosphoproteomics. Proteomics. 2015;15:340–55.
Zhang X, Maity T, Kashyap MK, Bansal M, Venugopalan A, Singh S, et al. Quantitative tyrosine phosphoproteomics of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor-treated lung adenocarcinoma cells reveals potential novel biomarkers of therapeutic response. Mol Cell Proteom. 2017;16:891–910.
Zhang X, Maity TK, Ross KE, Qi Y, Cultraro CM, Gao S, et al. Global proteome and phosphoproteome alterations in third generation EGFR TKI resistance reveal drug targets to circumvent resistance. bioRxiv 2020. https://doi.org/10.1101/2020.07.04.187617.
Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203.
Awasthi S, Maity T, Oyler BL, Zhang X, Goodlett DR, Guha U. Dataset describing the development, optimization and application of SRM/MRM based targeted proteomics strategy for quantification of potential biomarkers of EGFR TKI sensitivity. Data Brief. 2018;19:424–36.
Awasthi S, Maity T, Oyler BL, Qi Y, Zhang X, Goodlett DR, et al. Quantitative targeted proteomic analysis of potential markers of tyrosine kinase inhibitor (TKI) sensitivity in EGFR mutated lung adenocarcinoma. J Proteom. 2018;189:48–59.
Aoh QL, Castle AM, Hubbard CH, Katsumata O, Castle JD. SCAMP3 negatively regulates epidermal growth factor receptor degradation and promotes receptor recycling. Mol Biol Cell. 2009;20:1816–32.
Castle A, Castle D. Ubiquitously expressed secretory carrier membrane proteins (SCAMPs) 1-4 mark different pathways and exhibit limited constitutive trafficking to and from the cell surface. J Cell Sci. 2005;118:3769–80.
Thomas P, Wohlford D, Aoh QL. SCAMP 3 is a novel regulator of endosomal morphology and composition. Biochem Biophys Res Commun. 2016;478:1028–34.
Wu TT, Castle JD. Tyrosine phosphorylation of selected secretory carrier membrane proteins, SCAMP1 and SCAMP3, and association with the EGF receptor. Mol Biol Cell. 1998;9:1661–74.
Falguieres T, Castle D, Gruenberg J. Regulation of the MVB pathway by SCAMP3. Traffic. 2012;13:131–42.
Beaumatin F, O’Prey J, Barthet VJA, Zunino B, Parvy JP, Bachmann AM, et al. mTORC1 activation requires DRAM-1 by facilitating lysosomal amino acid efflux. Mol Cell. 2019;76:163–76.e8.
Tabara K, Kanda R, Sonoda K, Kubo T, Murakami Y, Kawahara A, et al. Loss of activating EGFR mutant gene contributes to acquired resistance to EGFR tyrosine kinase inhibitors in lung cancer cells. PLoS ONE. 2012;7:e41017.
Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–56.
Singleton DR, Wu TT, Castle JD. Three mammalian SCAMPs (secretory carrier membrane proteins) are highly related products of distinct genes having similar subcellular distributions. J Cell Sci. 1997;110:2099–107.
Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378:113–25.
Ghosh D, Ulasov IV, Chen L, Harkins LE, Wallenborg K, Hothi P, et al. TGFbeta-responsive HMOX1 expression is associated with stemness and invasion in glioblastoma multiforme. Stem Cells. 2016;34:2276–89.
Suarez-Arroyo IJ, Feliz-Mosquea YR, Perez-Laspiur J, Arju R, Giashuddin S, Maldonado-Martinez G, et al. The proteome signature of the inflammatory breast cancer plasma membrane identifies novel molecular markers of disease. Am J Cancer Res. 2016;6:1720–40.
Zhang X, Sheng J, Zhang Y, Tian Y, Zhu J, Luo N, et al. Overexpression of SCAMP3 is an indicator of poor prognosis in hepatocellular carcinoma. Oncotarget. 2017;8:109247–57.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Schiel JA, Simon GC, Zaharris C, Weisz J, Castle D, Wu CC, et al. FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nat Cell Biol. 2012;14:1068–78.
Diaz-Carballo D, Saka S, Klein J, Rennkamp T, Acikelli AH, Malak S, et al. A distinct oncogenerative multinucleated cancer cell serves as a source of stemness and tumor heterogeneity. Cancer Res. 2018;78:2318–31.
Mirzayans R, Andrais B, Murray D. Roles of polyploid/multinucleated giant cancer cells in metastasis and disease relapse following anticancer treatment. Cancers. 2018;10:118.
Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12:104–17.
Tomas A, Futter CE, Eden ER. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014;24:26–34.
Walsh AM, Lazzara MJ. Differential parsing of EGFR endocytic flux among parallel internalization pathways in lung cancer cells with EGFR-activating mutations. Integr Biol. 2014;6:312–23.
Padron D, Sato M, Shay JW, Gazdar AF, Minna JD, Roth MG. Epidermal growth factor receptors with tyrosine kinase domain mutations exhibit reduced Cbl association, poor ubiquitylation, and down-regulation but are efficiently internalized. Cancer Res. 2007;67:7695–702.
Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–6.
Glubb DM, Johnatty SE, Quinn MCJ, O’Mara TA, Tyrer JP, Gao B, et al. Analyses of germline variants associated with ovarian cancer survival identify functional candidates at the 1q22 and 19p12 outcome loci. Oncotarget. 2017;8:64670–84.
Kulyte A, Kwok KHM, de Hoon M, Carninci P, Hayashizaki Y, Arner P, et al. MicroRNA-27a/b-3p and PPARG regulate SCAMP3 through a feed-forward loop during adipogenesis. Sci Rep. 2019;9:13891.
Pinilla-Macua I, Grassart A, Duvvuri U, Watkins SC, Sorkin A. EGF receptor signaling, phosphorylation, ubiquitylation and endocytosis in tumors in vivo. Elife. 2017;6:e31993.
Pinilla-Macua I, Watkins SC, Sorkin A. Endocytosis separates EGF receptors from endogenous fluorescently labeled HRas and diminishes receptor signaling to MAP kinases in endosomes. Proc Natl Acad Sci U S A. 2016;113:2122–7.
Mittal K, Donthamsetty S, Kaur R, Yang C, Gupta MV, Reid MD, et al. Multinucleated polyploidy drives resistance to Docetaxel chemotherapy in prostate cancer. Br J Cancer. 2017;116:1186–94.
Parekh A, Das S, Parida S, Das CK, Dutta D, Mallick SK, et al. Multi-nucleated cells use ROS to induce breast cancer chemo-resistance in vitro and in vivo. Oncogene. 2018;37:4546–61.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.
Venugopalan A, Lee MJ, Niu G, Medina-Echeverz J, Tomita Y, Lizak MJ, et al. EGFR-targeted therapy results in dramatic early lung tumor regression accompanied by imaging response and immune infiltration in EGFR mutant transgenic mouse models. Oncotarget. 2016;7:54137–56.
Acknowledgements
We thank Gaga Geneti for the assistance with the mice handling and treatments. This research was supported by the NIH Intramural Research Program, Center of Cancer Research, National Cancer Institute.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
UG has a clinical trial agreement (CTA) with AstraZeneca and received research funding from AstraZeneca, Esanex and Aurigene. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Venugopalan, A., Lynberg, M., Cultraro, C.M. et al. SCAMP3 is a mutant EGFR phosphorylation target and a tumor suppressor in lung adenocarcinoma. Oncogene 40, 3331–3346 (2021). https://doi.org/10.1038/s41388-021-01764-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01764-y
This article is cited by
-
Homeobox D9 drives the malignant phenotypes and enhances the Programmed death ligand-1 expression in non-small cell lung cancer cells via binding to Angiopoietin-2 promoter
World Journal of Surgical Oncology (2023)
-
Enhancing the therapeutic efficacy of nanoparticles for cancer treatment using versatile targeted strategies
Journal of Hematology & Oncology (2022)
-
SCAMP2/5 as diagnostic and prognostic markers for acute myeloid leukemia
Scientific Reports (2021)