Abstract
HER3 is highly expressed in luminal breast cancer subtypes. Its activation by NRG1 promotes activation of AKT and ERK1/2, contributing to tumour progression and therapy resistance. HER3-targeting agents that block this activation, are currently under phase 1/2 clinical studies, and although they have shown favorable tolerability, their activity as a single agent has proven to be limited. Here we show that phosphorylation and activation of HER3 in luminal breast cancer cells occurs in a paracrine manner and is mediated by NRG1 expressed by cancer-associated fibroblasts (CAFs). Moreover, we uncover a HER3-independent NRG1 signaling in CAFs that results in the induction of a strong migratory and pro-fibrotic phenotype, describing a subtype of CAFs with elevated expression of NRG1 and an associated transcriptomic profile that determines their functional properties. Finally, we identified Hyaluronan Synthase 2 (HAS2), a targetable molecule strongly correlated with NRG1, as an attractive player supporting NRG1 signaling in CAFs.
Similar content being viewed by others
Introduction
Breast cancer is the leading cause of cancer-related mortality worldwide in females [1]. It is considered a heterogeneous disease that comprises several molecular subtypes based on gene expression analysis or biomarker expression [2, 3]. The family of human epidermal growth factor receptor (HER) of tyrosine kinases (TK) has four members, HER1/EGFR, HER2, HER3 and HER4, and eleven ligands [4]. Overexpression of HER family members favors cancer development, however, it also renders these tumours suitable targets for efficient anticancer therapies [5]. For instance, monoclonal antibodies (mAbs) such as trastuzumab and pertuzumab are usually employed in HER2 overexpressing subtypes [6, 7].
HER3 is emerging as an important component in the luminal subtype of breast cancer, which accounts for about 65–70% of all breast tumours [8]. In agreement with the observation that HER3 is required for cell survival in the luminal but not the basal mammary epithelium [9], luminal breast tumours present the highest levels of HER3 mRNA [10, 11]. HER3 has weak intrinsic TK activity and needs to form heterodimers with kinase-proficient receptor TKs to be functional [12]. For HER3-positive solid tumours, several HER3-targeting agents have been undergoing clinical evaluation for the last 10 years and currently thirteen mAbs are in phase 1 or 2 clinical studies. In contrast to HER2 inhibitors, HER3 binding antibodies such as lumretuzumab have shown limited clinical efficacy as single agents, but favorable tolerability [13, 14].
The major activating ligand of HER3 is neuregulin 1 (NRG1). Neuregulins (NRGs) are a family of the Epidermal Growth Factor (EGF) ligands that are widely expresed in solid tumors. Most isoforms are synthetized as a transmembrane pro-proteins that undergo proteolity cleavage liberating the EGF-like domain in the extracellular space [15]. In the presence of NRG1, HER3 heterodimerizes mainly with HER2, but also with EGFR or HER4 [16, 17]. These partner molecules induce HER3 tyrosine phosphorylation, binding of adapter molecules and thereby enabling downstream oncogenic signaling via PI3K/AKT, but also MAPK and JAK/STAT pathways. This ultimately leads to tumour progression [17, 18].
Several lines of evidence indicate that NRG1 contributes to the development and progression of different tumour types and its expression has been correlated with poor prognosis in breast cancer, head and neck squamous cell carcinoma and pancreatic cancer [19,20,21,22]. The fact that NRG1 is the main activating ligand of HER3, suggests that tumours with high levels of NRG1 could respond better to anti-HER3 targeted therapies [23,24,25]. Indeed, NRG1-autocrine signaling has been described in a subset of human cancers, such as head and neck and melanoma, to predict sensitivity to HER2/HER3 kinase inhibition [26, 27]. In the case of breast cancer, the relevance of NRG1 ligand in mediating resistance has been previously described [28]. However, in comparison to other cancer entities, the expression of NRG1 in breast tumour cells is usually low and the gene is frequently silenced by DNA methylation [29]. This suggests that an autocrine signaling is unlikely in breast cancer and rather the activation of HER3 in luminal cancer cells might be dependent on NRG1 expressed by cells in the tumour microenvironment.
The tumour microenvironment is typically composed mainly of cancer-associated fibroblasts (CAFs) acompained by immune cells, vascular cells and extracellular matrix (ECM) [30]. CAFs are characterized by the expression of activation markers such as αSMA (alpha smooth muscle actin), FAP (fibroblast activation protein) and FSP1 (fibroblast-specific protein 1) [31], and are a known source of ECM and soluble factors (e.g. growth and inflammatory factors) which impact tumour growth and progression. The potential of CAFs as therapeutic targets or prognostic biomarkers is still under debate, as CAFs appear to represent a heterogeneous group of cells with diverse and even opposing functions that differentially determine tumour fate [32,33,34].
Here, we study CAF heterogeneity in luminal breast cancer both at the molecular and functional level. Using primary CAFs derived from tumour tissue of luminal breast cancer patients, we demonstrate how heterogeneous expression of NRG1 in CAFs determines response of cancer cells to therapies blocking the HER3 signaling pathway [35, 36]. In addition, we uncover an HER3-independent role of NRG1 associated with migration and proliferation of CAFs, and identified a NRG1-correlating transcriptomic network enriched in motility and fibrosis present in CAFs. Finally, we reveal Hyaluronan Synthase 2 (HAS2), a targetable molecule, as a supporting player strongly correlating with NRG1 expression in primary fibroblasts and patient data.
Results
NRG1 is expressed in the stromal compartment of luminal breast cancer
To verify the expression pattern of HER3 in different breast cancer subtypes, we used the public METABRIC [37] and TCGA [38] gene expression datasets. In accordance with previous reports [9], HER3 showed consistent higher expression in the luminal subtypes in both datasets (Fig. S1A). Conversely, the expression of its main ligand NRG1 was overall lower with higher levels in basal-like subtypes (Fig. 1A).
A Dot plots representing expression of NRG1 in breast cancer subtypes (PAM50) extracted from METABRIC [37] and TCGA [38] datasets. Mean + /− s.e.m represented. Only statistically significant comparisons with luminal subtypes are depicted. ANOVA multiple comparison test (***P < 0.001). B Dot plots representing expression of NRG1 in the epithelial and stromal compartment in four laser capture microdissected (LCM) breast cancer datasets (GSE10797, n = 28; GSE14548, n = 14; GSE35019, n = 53; and GSE83591, n = 39). Mean + /− s.e.m represented. Two-tailed paired Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001). C Viability of T47D (black) and MCF7 (grey) measured 72 h after treatment with HER3 mAb lumretuzumab or pertuzumab at indicated doses. Values represent median of three independent experiments (n = 5 technical replicates). U-Mann–Whitney two-tailed test was applied. No significant differences were observed. D Ectopic NRG-1β (50 ng/mL) was added to T47D and MCF7 cell lines and viability quantified. Cells treated with DMEM–F12 media containing 1% FCS were used as a control. Results conditions where cells had been preincubated for 1 h with lumretuzumab (Lum) or pertuzumab (Pert) at 10 µg/mL were normalized to the control condition without NRG1. Two-tailed unpaired Student’s t-test (*P < 0.05; **P < 0.01) comparing each treatment with untreated condition (with NRG1). Bars represent average of two independent experiments + /− s.e.m. E Western blot showing HER3 and downstream effectors AKT and ERK1/2 (total and phosphorylated levels), 5 min after addition of NRG-1β (50 ng/ml). Some samples were either pre-incubated with mAbs lumretuzumab (Lum) or pertuzumab (Pert) at 10 µg/ml for 1 h. Images are representative of three biological replicates.
Gene expression analysis of bulk tissues comprises mixed signals from different cellular components, masking the contribution of different tumour compartments. Thus, we next explored NRG1 expression in a collection of breast cancer datasets generated by laser capture microdissection (LCM) of the stromal and epithelial compartments (GSE10797; [39], GSE14548; [40], GSE35019 [41] and GSE83591 [42]). In all LCM datasets explored, expression of NRG1 was higher in the stromal compartment (Fig. 1B). This indicates that the stromal cells are the major contributors of NRG1 expression in breast tumour tissue and suggests that activation of the HER3 pathway in tumour cells preferentially happens in a paracrine manner.
In order to define a proper in vitro system for subsequent studies, we analyzed different breast cancer cell lines for expression of the HER family receptors (EGFR, HER2, HER3 and HER4). As in the primary tissue datasets, cancer cell lines from luminal subtypes (T47D, MCF7 and BT474) showed elevated levels of HER3 (Fig. S1B).
We focused on luminal A cell lines T47D and MCF7 to avoid masking of HER3 mediated effects by HER2 overexpression (BT474). To test if luminal A cancer cell lines might be intrinsically addicted to HER3 oncogenic signaling [43], cells were challenged with increasing doses of the therapeutic monoclonal antibody lumretuzumab, which blocks binding of NRG1 to HER3 [44], or pertuzumab, which blocks HER2/HER3 heterodimer formation [45]. After 3 days of treatment, viability of cancer cell lines was not affected by HER3 blockage, suggesting no autocrine activation of the HER3 pathway in the luminal A cell lines (Fig. 1C). However, HER3 might still be relevant via paracrine activation. To test this, we added ectopic NRG1 to cancer cells that had or had not been pre-incubated with either lumretuzumab or pertuzumab. Whereas the viability of control cells without antibody-incubation was indeed increased by ectopic NRG1, the effect was abolished by pre-treatment with lumretuzumab or pertuzumab (Fig. 1D). In addition, phosphorylation of HER3 and of its main downstream effectors AKT and ERK was efficiently induced by NRG1 in control cells, while this was strongly prevented upon pre-treatment with lumretuzumab or pertuzumab (Fig. 1E and S1C, D). Together, these data demonstrate that NRG1 activates HER3 pathway via binding to HER3 in a paracrine manner and that this paracrine activation can be blocked with monoclonal antibodies.
Primary breast cancer-associated fibroblasts express variable levels of NRG1
Cancer-associated fibroblasts (CAFs) are particularly abundant in the stroma of solid tumours [46]. As we had observed that NRG1 is highly expressed in the stromal compartment of breast tumour tissue (Fig. 1B), we next aimed to determine if CAFs are a source of NRG1. To this end, we established primary cultures of CAFs derived from tumour tissues from six breast cancer patients clinically classified as luminal subtype (Table S1).
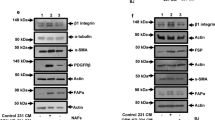
The isolated cells showed the characteristic fibroblast morphology as well as expression of common CAF activation markers such as αSMA, fibronectin (FN1) and FAP (Fig. 2A–C and S2A). We further confirmed their CAFs lineage by comparing RNA seq expression data of specific markers in CAFs vs cancer cell lines (Table S2).
A, B Immunofluorescence (A) and western blot (B) of common activation markers αSMA (red, upper panel) and fibronectin (FN1) (red, lower panel) in CAFs and luminal cancer cells lines (T47D and MCF7) under study. In (A), nuclei counterstaining with DAPI (blue). Representative images are shown. In (B), the epithelial marker E-cadherin served as marker for epithelial cells. Tubulin was used as a loading control. C Heatmap representing protein values of common mesenchymal markers αSMA, fibronectin, vimentin and FAPα in all six CAFs under study and in two luminal breast cancer cell lines obtained by reverse phase protein array (RPPA). Color intensities are ranked per each antibody (red = maximum, blue = minimum). D Expression of NRG1 transcript in all six CAFs and in luminal breast cancer cell lines (T47D and MCF7). Values were normalized to the geometric mean of ACTB and GAPDH, and are shown as relative to CAF#4. Bars represent mean + /− s.e.m of two independent experiments.
Next, we analysed mRNA transcript and protein expression levels of NRG1 in the CAF lines and in the two luminal breast cancer cell lines. In line with the LCM patient data, NRG1 was expressed by all CAF lines, while no expression could be detected in cancer cells (Fig. 2D and S2B). Interestingly, NRG1 levels were heterogeneous among the different fibroblasts despite having been isolated from tumours of the same subtype. Collectively, these results demonstrate that NRG1 is expressed by CAFs in the stroma of luminal breast cancer patients and reinforce the concept of a paracrine-driven activation of HER3 in the luminal breast cancer subtype.
Different levels of NRG1 secreted by CAFs determine activation of HER3 in cancer cells
To ascertain whether different expression of NRG1 by CAFs translates into variable activation of the HER3 pathway in cancer cells, we stimulated T47D and MCF7 cancer cells with conditioned media (CM) from the isolated CAFs, and used lumretuzumab to block ligand–receptor binding.
In order to detect phosphorylation levels of HER3 and its main downstream effectors AKT and ERK in a sensitive and quantitative manner, we applied Reverse Phase Protein Array (RPPA) technology [47]. Incubation with ectopic NRG1 as well as blockage with lumretuzumab were used as positive controls (Fig. S3A). Phosphorylation of HER3 observed in cancer cells upon stimulation with the different CM was CAF- and cancer cell-dependent, achieving different phosphorylation levels (Fig. 3A, red = maximum, blue = minimum). In both cell lines we observed variable levels of phosphorylation of the HER3 pathway induced by CM from the different CAFs, which was specially stronger for CAF#3, the one that showed higher expression of NRG1 (Fig. 2D and Fig. S2B). Pre-incubation with lumretuzumab reduced phosphorylation of HER3 and its effectors in all conditions, confirming a HER3 activation mediated by the NRG1-present in the CAF-CM.
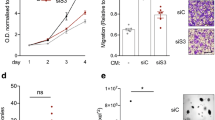
A Heatmap representing relative phosphorylation levels of HER3, AKT and ERK1/2 in T47D and MCF7 cancer cells, quantified by RPPA. CAF conditioned media (CM) was added to cancer cells for 5 min that had or had not been pre-incubated with lumretuzumab (Lum) (10 µg/ml) for 1 h. All CM were obtained with 1% FCS, and DMEM-F12 with 1% FCS was used as control. Values represent median of three technical replicates and three biological replicates. Color intensities are ranked per each antibody (red = maximum, blue = minimum). B Relative proliferation of T47D and MCF7 cancer cells after 72 h with different CAF-CM with lumretuzumab (red squares) or untreated (black circles), compared with cells cultured in control media (DMEM-F12, 1% FCS) and untreated. Dots/squares represent mean + /− s.e.m of three independent biological replicates (n = 4). P values were determined by two-tailed U-Mann–Whitney test for each CM (*P < 0.01; **P < 0.001). C Percentage of closure in a scratch assay of T47D or MCF7 cancer cells, after 21 h of treatment with 10 μg/ml lumretuzumab (red) or untreated (black), and with conditioned media (CM) of indicated CAFs. DMEM-F12 1% FCS was used as negative control and NRG-1β (50 ng/ml) as positive control. Box plots correspond to the mean and + /− s.e.m. of two independent experiments (each with n = 6 technical replicates). Two-tailed U-Mann–Whitney test for each CM (*P < 0.01; **P < 0.001; ***P < 0.0005; ****P < 0.0001).
Next, we tested the ability of NRG1 in the CAF-CM to promote proliferation of cancer cells. The proliferation rate of cancer cells did not follow the same trend as NRG1 expression; however, blockage of NRG1-HER3 signaling by lumretuzumab, decreased proliferation (Fig. 3B) of T47D cancer cells while a minor effect was observed for MCF7 cells.
Due to the established role of NRG1 in epithelial to mesenchymal transition (EMT) and migration processes [48], we next measured if also migration abilities of the cancer cells were altered in presence of CAF-CM and if these were dependent on HER3 activation by NRG1. Consistently, CAF-CM increased migration of MCF7 cancer cells mainly, and the effect was diminished with lumretuzumab whereas a less prominent effect was obtained for T47D cancer cells. (Fig. 3C). The strongest effect was observed for CAF#3-CM, the CAF culture that expressed the highest level of NRG1.Treament with lumretuzumab of cancer cells cultured in control media, did not alter their migration abilities, confirming the absence of an autocrine HER3 activation (Fig. S3B). While the T47D cell line showed a boosting effect in proliferation but more modest migration phenotype, the opposite occurred for the MCF7 cell line. The latter showed little or non-increase in proliferation while stronger effects were observed for migration, which could be explained by the molecular switch from proliferative to migratory phenotype, where cells reduce their proliferation rate in order to enhance their migratory potential.
Taken together, these results indicate that CAFs isolated from tumour tissue of luminal breast cancer specimens differently activate the HER3 pathway and regulate proliferation and/or migration of cancer cells via NRG1 secretion.
Heterogeneous expression of NRG1 among CAFs
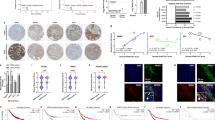
Despite all different cultures of CAFs were derived from luminal breast cancer tissue, they showed variable capacities to activate the HER3 pathway in luminal breast cancer cells via NRG1 (Fig. 3A–C). To investigate the possible differences between the isolated CAFs in a global approach, we performed RNA sequencing of the six primary CAF lines. Principal component analysis (PCA) revealed that gene expression among fibroblasts was indeed scattering (Fig. 4A). To elucidate which genes were contributing to this variance, we analyzed the most significant variable genes (MVG) amongst the different CAF lines. A list of 517 significant genes was defined, with NRG1 ranked among them (Table S3). Other genes listed as highly heterogeneous were ACTA2 and S100A4 coding for αSMA and FSP1 respectively, two well accepted CAF markers, although not correlated with NRG1 expression (Fig. S4A).
A Principal component analysis PC1 and PC2 of all six CAF lines under study. B Biological processes (gene ontology) enriched in CAFs most variable genes (MVG). Graph bars represent adjusted P value. C Proteomic profile of CAFs based on reverse phase protein array (RPPA). Intensity values (red = maximum, blue = minimum) are ranked per each antibody to compare between samples. Values represent median of three technical replicates. Unsupervised clustering by Euclidian distance separates CAFs (high-NRG1, blue and low-NRG1, grey) respectively.
Next, we performed functional analysis using gene ontology (GO) terms in the Bioinfominer online platform (https://bioinfominer.com) [49] for the 517 MVG. Functional categories related to extracellular matrix, cell adhesion and locomotion were significantly enriched (Fig. 4B and Table S4). Among the functional categories that were differing in fibroblasts, we found regulation of proliferation. In line with this, nuclei counting of fibroblasts along several days revealed a heterogeneous proliferation degree among the CAFs lines (Fig. S4B).
To comprehend upto which extent NRG1 contributed to fibroblasts variability, we split CAFs based on NRG1 expression into low-NRG1 (lower than mean: CAF#1, CAF#4, CAF#5) and high-NRG1 (higher than mean: CAF#2, CAF#3, CAF#6) CAFs (Fig. S4C). We perfomed targeted proteomic analysis for the CAFs under study to elucidate if phosphorylation status of effectors of the HER pathway (e.g. ERK1/2, AKT, MET, S6K, ADAM17 [50]) were differing among the lines. Unsupervised clustering grouped CAFs into two groups demonstrating different activation of the HER3 pathway. Interestingly, NRG1 expression was sufficient to separate those CAFs (Fig. 4C). Moreover, analysis of the transcription factor genes enriched in high-NRG1 CAFs disclosed c-JUN as the main transcription factor regulating high-NRG1 transcriptome (Fig. S4D and Table S4 Table S5). Strikingly, phosphorylation of the transcription factor c-JUN, described as a central molecular mediator in fibrotic conditions [51] and hyperactivated in high density stroma breast cancer tissue [52], was higher in high-NRG1 CAFs (CAF#2, CAF#3, CAF#6).
Collectively, these results underline the relevance of NRG1 as a heterogeneous factor expressed by CAFs in luminal breast tumours.
NRG1-associated transcriptome correlates with migration processes in CAFs
We next wanted to elucidate if different expression of NRG1 in fibroblasts was associated with specific transcriptional programs. To this end, we performed differential expression analysis between the designated high and low-NRG1 groups of CAFs. A total of 102 genes were upregulated and 151 were downregulated in the high- vs low-NRG1 CAFs (adj P value < 0.05 and absolute logFC > 0.5) (Table S6). Gene Ontology (GO) analysis revealed that genes enriched in the high-NRG1 CAF group were mainly related with adhesion and motility processes. In contrast, terms enriched in low-NRG1 CAFs were associated to signaling and metabolic processes (Fig. 5A). Based on the list of significant differentially expressed genes (Table S6), we used BioinfoMiner (https://bioinfominer.com) to explore systemic processes and driver genes characteristic of each group. In line with the GO results (Fig. 5A), driver genes (P value < 0.002 and log2FC > 2) in high-NRG1 CAFs included ITGB2, EPHB1 and HAS2, known locomotion and extracellular matrix reorganization related genes. In contrast, driver genes in low-NRG1 CAF included genes such as PTGIS, TRH, WNT2 or JAG1, involved in cell signaling and metabolic processes (Table 1).
A Functional classification of genes by gene ontology (GO). Biological processes represented in each group of CAFs. Graph bars showing number of genes with significant P value for high- (blue) and low- (grey) NRG1 CAFs respectively. B Expression correlation of NRG1 and candidate genes quantified by RT-PCR in primary CAFs from two independent sources, six lines from Halle University (discovery set, blue = high-NRG1 and grey = low-NRG1) and another nine from Breast Cancer Now (validation set, black). Each dot represents the average of >3 independent experiments. Spearman r correlation and two-tailed P value are indicated. C Expression correlation of NRG1 and indicated genes in the stroma compartment of the LCM dataset GSE83591 (n = 39). Spearman correlation r and P values are indicated.
Altogether, these results determine NRG1 as a stromal marker discerning fibroblasts with different transcriptional programs.
HAS2 expression correlates with NRG1 in the tumour stroma of patient samples
In order to obtain a signature of genes linked with NRG1 expression in tumour stromal fibroblasts, we selected those genes with the strongest correlation with NRG1 (Pearson r > 0.8), and with a log2FC > 2 in high- vs low-NRG1 CAFs (Fig. S5A and Table S7).
We validated the correlation of these genes (i.e. ITGB2, EPHB1 and HAS2) with NRG1 in a second set of primary CAFs from an independent source (https://breastcancernow.org/breast-cancer-research/breast-cancer-now-tissue-bank) (Fig. 5B), and we explored their expression in LCM stroma datasets from breast cancer patients (Fig. 5C). This analysis confirmed a significant positive correlation between NRG1 and HAS2 (Fig. 5C). Next, we checked the expression in the TCGA dataset covering bulk tumour tissue, only considering those samples with tumour purities <50% to select tumours with high stroma content. From the candidates investigated, HAS2 was significantly correlated with NRG1 also in TCGA dataset (Fig. S5B).
Taken together, these analyses uncover HAS2 as a stromal gene highly correlated with NRG1 in luminal breast cancer patients.
NRG1 downregulation in CAFs downregulates HAS2 and impairs their migration
We had observed that high-NRG1 CAFs displayed higher proliferation rates and showed signatures of proliferation (Fig. S4B and Table S4). We thus explored the potential contribution of NRG1 expression to this phenotype in CAFs. To this end, we first knocked down NRG1 in high-NRG1 CAF lines by transient transfection. We used two independent siRNA sequences inducing different degrees of downregulation, thus mimicking heterogeneous downregulation of NRG1 at RNA and at protein level, both secreted and cell-bounded (Fig. 6A, B and Fig. S6A). Functional downregulation of NRG1 was confirmed by using the CAF-siNRG1 CM on cancer cells. Both T47D and MCF7, showed a decrease in HER3 phosphorylation after incubation with CM from siNRG1 transfected CAF#3, which paralleled the extent of NRG1 downregulation in the CAFs (Fig. S6B). Similarly, migration (Fig. S6C) and proliferation of cancer cells induced by CAFs CM was also diminished upon NRG1 downregulation in CAF#3. Effects obtained with siNRG1#3 were comparable to those obtained upon lumretuzumab treatment (Fig. S6D). Similar pattern was obtained with the CM of CAF#2 and CAF#6, although showing a less prominent effect (Fig. S6E) probably due to the different basal level of NRG1 in the control condition. Having shown that the levels of NRG1 downregulation in CAFs were sufficient to affect cancer cells, we next investigated the effect of NRG1 in CAFs. We observed that decreased NRG1 expression resulted also in a reduced proliferation rate in CAFs themselves (Fig. 6C); however, ectopic addition of NRG1 did not rescue that phenotype (Sup Fig. S6F). This indicates that CAF-secreted NRG1 positively contributes to proliferation of cancer cells while it does not affect CAFs. Thus, we next investigated if the proliferative effect of NRG1 expression in CAFs was dependent on the binding of NRG1 to HER3. Contrary to cancer cells (Fig. 3B and Fig. S1D), inhibition of NRG1 binding to HER3 by lumretuzumab, neither decrease proliferation of CAFs nor phosphorylation of AKT and ERK (Fig. 6D and E). Altogether, these data demonstrate that the effect exerted by NRG1 on CAFs proliferation is independent of the canonical binding of secreted NRG1 to HER3. Further supporting this finding, expression levels of HER3 in CAFs were very low compared with expression levels in cancer cells and even lower in high-NRG1 CAFs (Fig. S6G). Of note, binding of NRG1 to HER4 receptor was not considered due to its undetectable expression in CAFs (Fig. S6H).
A Downregulation of NRG1 at RNA level (A) in CAF#3 (high-NRG1) using two independent siRNAs (siNRG1 #1, #3). Bar graphs represent average of three independent experiments and three technical replicates each. Two-tailed U-Mann–Whitney test comparing with siRNA non-targeting control (siNTC) (***P < 0.01). B Downregulation of NRG at protein level using two independent siRNAs (siNRG1 #1, #3). Left panel shows secreted NRG1 in the conditioned media measured using ELISA assay. Right panel shows western blot of cell-bounded NRG1 (upper) and secreted NRG1 in conditioned media obtained via immunoprecipitation (lower), were concentrated DMEM was used as a negative control. Images are representative of two independent experiments. C Relative cell growth of CAF#3 was measured 72 h post re-seeding (5 days post transfection) either with two different siRNAs targeting NRG1 (siNRG1 #1, #3) or a non-targeting control siRNA (siNTC). U-Mann–Whitney test was applied for statistical analysis (***P < 0.01). D Relative cell number of CAF#3, 72 h after treatment with 10 μg/ml lumretuzumab (Lum). No statistical differences were obtained. E Total protein and phosphoprotein levels for AKT and ERK1/2 in CAF#3 after 24 h of treatment with or without lumretuzumab (Lum) at 10 μg/ml. GAPDH was used as loading control. F HAS2 mRNA levels in cells treated either with a non-targeting control siRNA (siNTC) or two different siRNAs targeting NRG1 (siNRG1#1, #3) relative to siNTC. Graph bars represent average of three independent experiments (n = 3 technical replicates). Two-tailed U-Mann–Whitney test comparing with siRNA non-targeting control (siNTC) (*P < 0.05; ***P < 0.001). G Migration of CAFs transfected either with a non-targeting control siRNA (siNTC) or two siRNAs targeting NRG1 (siNRG1#1, siNRG1#3) and normalized with seeding control. Whiskers in the box plot represent minimum and maximum values of three independent experiments (six technical replicates each). Two-tailed U-Mann–Whitney test comparing with siRNA non-targeting control (siNTC) (**P < 0.01). Crystal violet staining of transwell inserts representing migration after 8 h of CAF#3 transfected either with a control siRNA (siNTC) or either of siRNAs targeting NRG1 (siNRG#1, siNRG1#3).
Transcriptomic profiling had revealed an enrichment of signatures related to migration in high-NRG1 CAFs (Fig. 5A). In addition, NRG1 correlated with HAS2, a known mediator of migration, in several CAF lines and patient stroma datasets (Fig. 5B and C). We thus wondered if NRG1 downregulation could affect also HAS2 levels and migratory capacity of CAFs. Remarkably, we observed a proportional decrease in HAS2 mRNA transcript levels upon knockdown of NRG1 (Fig. 6F) which was associated with a marked decrease of the migration of CAFs (Fig. 6G).
Collectively, this data suggest a HER3-independent role of NRG1 in cancer-associated fibroblasts that modulates their proliferation and migration and that could be driven by its intracellular domain (ICD).
Discussion
Overexpression of human epidermal growth factor receptor 3 (HER3) plays an important role in cancer development as well as acquired drug resistance in a wide variety of solid tumours [53, 54]. It has been associated with worse clinical outcome, and monoclonal antibodies such as lumretuzumab have been developed to neutralize its activity by blocking the binding of its ligand NRG1 [35, 44]. Several pre-clinical and clinical studies have supported NRG1 as a predictive biomarker for anti-HER3 targeted therapies [13, 23, 55]. NRG1-mediated autocrine signaling in cancer cells has been reported to underlie sensitivity to anti-HER2 therapies in certain ovarian and head and neck tumours [26, 56]. In our study, we show that in luminal breast cancer, the stromal compartment is the major contributor of NRG1 expression and that its expresion is non detectable in cancer cells [29]. Our results suggest that in luminal breast cancer, cancer cells depend on paracrine NRG1 to activate downstream pathways. Here, we used primary fibroblasts isolated from luminal breast cancer tissue and demonstrated that NRG1 produced and secreted by CAFs is sufficient to activate the HER3 pathway in cancer cells. Activation of HER3 by CAF CM promotes phosphorylation of main downstream activators AKT and ERK, leading to proliferation and migration of cancer cells. The use of lumretuzumab, a humanized monoclonal antibody that selectively binds to the extracellular domain of HER3 thereby blocking the binding of NRG1, is able to prevent that phenotype in a NRG1-dependent manner, as similarly occurred when using of CM from NRG1 downregulated CAFs. Thus, we suggest that the utility of NRG1 as a predictive biomarker to anti-HER3 therapies in luminal breast cancer may be provided by the stromal compartment, while analysis of bulk tumour tissues may dilute its detection [57].
It is widely accepted that CAFs are a heterogeneous population of mesenchymal cells defined by their diversity in functions, markers and origins [34]. Several studies have compared gene expression in disease-free fibroblasts and CAFs derived from various tissues to obtain information on stromal pathways facilitating malignant phenotypes [58,59,60]. Other works have been oriented towards identifying specific lineages within CAFs based on their tumour promoting abilities to identify subpopulations [61,62,63]. Also, recent studies have described novel approaches for the study of biological function and targeting of CAFs [64]. Here, we have identified heterogeneous expression of NRG1 in the stroma of luminal breast cancer tissue. Its higher expression defines CAFs with an associated motile, fibrotic transcriptome and phenotype. In addition, unsupervised clustering based on the proteomic profile of relevant signaling effectors such as ERK1/2, AKT,MEK, and ADAM17 (a disintegrin metalloprotease responsible for the cleavage of membrane bound growth factors such as NRG1 [65]), classified CAF lines in the same high- and low-groups, supporting the role of NRG1 in defining a different activation status.
The differential expression analysis conducted in this study revealed 102 genes upregulated in high-NRG1 CAFs, which were enriched in gene signatures related to a motile phenotype. We identified ITGB2 and EPHB1 as strongly correlating with NRG1 expression in breast CAFs. Indeed, both ITGB2 and EPBH1 have been previously documented to play significant roles in polarization and cell migration [66].
Finally, we revealed HAS2 (Hyaluronan Synthase 2) as a gene that is strongly correlated with NRG1, not only in primary CAFs but also in patient stroma datasets. HAS2 is responsible for the synthesis of hyaluronan (HA), a glycosaminoglycan with a demonstrated role in cancer initiation and progression and whose elevated accumulation in either the stroma or tumour parenchyma of many cancers is linked to tumour aggressiveness and poor outcome [67, 68]. We suggest that in our system, correlation of HAS2 and NRG1 is consequence of a regulatory mechanism in which NRG1 expressed by CAFs regulates HAS2 expression and modulates migratory potential of the fibroblasts by NRG1 non-canonical signaling [69, 70]. We hypothesize that this regulation is the result of NRG1 backward signaling, which has been previously described in the development of the nervous system [71]. We suggest that proteolytic cleavage of pro-NRG1 in the intracellular domain induces the release of NRG1 ICD, which triggers backward signaling by nuclear translocation and modulation of gene transcription. Although additional molecular characterization will be necessary to further decipher the exact mechanism of this regulation, the strong correlation observed in tumour stroma of luminal breast cancer patient samples clearly propose these two molecules as potential CAFs biomarkers and therapeutic targets. Thus, we consider that dual targeting of NRG1 and HAS2 may be an interesting treatment strategy applicable for tumours with high expression of NRG1. Indeed, inhibition of HAS2 upon treatment with its specific inhibitor 4-methylumbelliferone (4-MU, also known as hymecromone), has proven successful in reducing tumour stroma in pancreatic ductal adenocarcinoma (PDAC) [72, 73], which also shows high expression of NRG1 [24]. We hypothesize that for breast cancer, on one side, treatment with anti-HER3 monoclonal antibodies would reduce the proliferation and migration of cancer cells by blocking stromal NRG1 binding, thus diminishing tumour aggressiveness. Concomitantly, the use of a specific HAS2 inhibitor would induce a reduction of the stroma content by decreasing HA synthesis by CAFs (Fig. 7) further contibuting to tumour growth/aggressiveness wane. Notably, both lumretuzumab and 4-MU are approved drugs currently used in several clinical trials. Ultimately, this tailored combination therapy represents a novel treatment approach from which not only luminal breast cancer patients but possibly other tumor entities with high levels of stromal NRG1 could benefit.
High-NRG1 CAFs (blue) secrete high quantities of NRG1 (blue circles) promoting paracrine activation of HER3-receptor in cancer cells and thereby inducing proliferation and migration processes. Use of lumretuzumab blocks HER3 receptor in cancer cells and avoids binding of NRG1 thereby reducing proliferation and migration processes in the tumour cells. Suggested NRG1 non-canonical signaling induces proliferation and migration in CAFs, in a HER3-independent manner. CAFs are highly proliferative and migratory also due to increased expression of HAS2 and secretion of HA. Additional use of a HAS2 inhibitor could help to reduce expression of HAS2, and thus secretion of HA reducing migration of CAFs (/https://smart.servier.com/).
Materials and methods
Clinical samples, cell isolation and characterization
Tumour tissue was collected from patients (n = 6) undergoing surgery for breast carcinoma at the Department of Gynecology, Martin-Luther-University Halle-Wittenberg in Halle (Saale), Germany, following ethical approval by the Ethics committee of Martin-Luther-University Clinics Halle-Wittenberg (Halle Saale), Germany, and written informed consent provided by the patients. Tumour tissue was mechanically minced into pieces (1–4 mm3) and centrifuged at 1600 rpm for 10 min. After fat removal, pellet containing small pieces was resupended in DMEM/F12 (10% FCS, 1% P/S and 1% fungizone), filtered with a cell strainer (70 µm) and plated in a 60 mm culture dish. Outgrowth of cells was daily checked and medium renewed twice per week. After complete outgrowth in a 60 cm2 dish, cells were passaged with 0.25% Trypsin-EDTA (Gibco, Life Technologies) and fibroblasts seeded into a new 100 mm culture dish. After three cell passages, morphologically homogeneous cultures containing only fibroblasts were obtained and RNA and protein were collected for further characterization. To obtain conditioned media, 2.5 × 105 CAFs were seeded in 100 mm culture dish. Once cells were attached, media was replaced by DMEM/F12 (1% FCS, 1% P/S), incubated for 24 h and collected for their further use.
Reverse Phase Protein Array (RPPA)
RPPA experiments were performed as previously described [47, 74]. Briefly, cell lysates from three biological replicates for each condition were spotted in nitrocellulose-coated glass slides (Oncyte Avid, Grace-Biolabs, Bend, OR, USA) in technical triplicates. All the primary antibodies used were previously validated through Western blots to test their specificity. Signal intensities of spots were quantified using GenePixPro 5.0 (Molecular Devices, Sunnyvale, CA, USA). Intensity values were log2 transformed and plotted using morpheus software (https://software.broadinstitute.org/morpheus/). List of antibodies used is provided in Table S8
Drug treatments
Lumretuzumab and pertuzumab were provided by Roche Diagnostics GmbH (Penzberg). Prior to addition of CAF-CM or human recombinant NRG-1β (4711, BioCat), cells were pre-treated with either lumretuzumab or pertuzumab (10 μg/ml) for 30 min in low serum media (1% FCS). For viability/proliferation assays, media was removed and CAF-CM or low serum media (with or without 50 ng/ml NRG-1β) was added and incubated for 3 days. For short perturbation assays, incubation time was 5 min prior to lysates collection.
RNA sequencing and data processing
RNA sequencing of the six CAF lines was performed at the Genomics Core Facility of German Cancer Research Center (DKFZ- https://www.dkfz.de/gpcf) using the Illumina HiSeq 4000 platform and the Illumina stranded TruSeqRNA paired end sequencing kit. After quality control, reads were mapped to the human genome hg38 using STAR (version 2.3.0e), and reverse strand read counts were determined using featureCounts (version 1.5.1). The reads were mapped to ENSEMBL IDs and gene symbols. Exonic gene lengths and TPMs (Transcript Per kilobase Million reads) were calculated in R. To avoid infinite values, a count of one was added to each TPM value and the resulting values were then log2 transformed.
Functional analysis: gene ontology
We used the BioInfoMiner online platform to investigate which biological process Gene Ontology (GO) terms [75, 76] were enriched in the list of differentially expressed or highly variable genes. BioInfoMiner exploits biological hierarchical vocabularies by mapping the genes to a genomic network created from semantic data. It prioritizes them based on the topological properties of the network after minimizing the impact of semantic noise (bias) through different types of statistical correction. It detects and ranks significantly altered processes and the driver genes involved. The BioInfoMiner platform is available online at the website https://bioinfominer.com.
Data availability
Gene expression data of the primary CAF lines is available in the ArrayExpress data repository at https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-10075. All other data generated or analysed during this study are included in this published article [and its supplementary information files].
References
Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends-an update. Cancer Epidemiol Biomark Prev. 2016;25:16–27.
Sorlie T. Molecular portraits of breast cancer: tumour subtypes as distinct disease entities. Eur J Cancer. 2004;40:2667–75.
Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ, et al. Strategies for subtypes-dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the primary therapy of early breast cancer 2011. Ann Oncol. 2011;22:1736–47.
Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34.
Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–54.
Roskoski R Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharm Res. 2014;79:34–74.
Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25:282–303.
Cejalvo JM, Martinez de Duenas E, Galvan P, Garcia-Recio S, Burgues Gasion O, Pare L, et al. Intrinsic subtypes and gene expression profiles in primary and metastatic breast cancer. Cancer Res. 2017;77:2213–21.
Balko JM, Miller TW, Morrison MM, Hutchinson K, Young C, Rinehart C, et al. The receptor tyrosine kinase ErbB3 maintains the balance between luminal and basal breast epithelium. Proc Natl Acad Sci USA. 2012;109:221–6.
Fujiwara S, Ibusuki M, Yamamoto S, Yamamoto Y, Iwase H. Association of ErbB1-4 expression in invasive breast cancer with clinicopathological characteristics and prognosis. Breast Cancer. 2014;21:472–81.
Morrison MM, Hutchinson K, Williams MM, Stanford JC, Balko JM, Young C, et al. ErbB3 downregulation enhances luminal breast tumor response to antiestrogens. J Clin Invest. 2013;123:4329–43.
Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci USA. 2010;107:7692–7.
Jacob W, James I, Hasmann M, Weisser M. Clinical development of HER3-targeting monoclonal antibodies: perils and progress. Cancer Treat Rev. 2018;68:111–23.
Meulendijks D, Jacob W, Martinez-Garcia M, Taus A, Lolkema MP, Voest EE, et al. First-in-human phase i study of lumretuzumab, a glycoengineered humanized anti-HER3 monoclonal antibody, in patients with metastatic or advanced HER3-POSITIVE SOLID Tumors. Clin Cancer Res. 2016;22:877–85.
Willem M. Proteolytic processing of Neuregulin-1. Brain Res Bull. 2016;126:178–82.
Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–16.
Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–37.
Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–67.
Montero JC, Rodriguez-Barrueco R, Ocana A, Diaz-Rodriguez E, Esparis-Ogando A, Pandiella A. Neuregulins and cancer. Clin Cancer Res. 2008;14:3237–41.
Qian G, Jiang N, Wang D, Newman S, Kim S, Chen Z, et al. Heregulin and HER3 are prognostic biomarkers in oropharyngeal squamous cell carcinoma. Cancer. 2015;121:3600–11.
Kolb A, Kleeff J, Arnold N, Giese NA, Giese T, Korc M, et al. Expression and differential signaling of heregulins in pancreatic cancer cells. Int J Cancer. 2007;120:514–23.
Tsai MS, Shamon-Taylor LA, Mehmi I, Tang CK, Lupu R. Blockage of heregulin expression inhibits tumorigenicity and metastasis of breast cancer. Oncogene. 2003;22:761–8.
Ocana A, Diez-Gonzalez L, Esparis-Ogando A, Montero JC, Amir E, Pandiella A. Neuregulin expression in solid tumors: prognostic value and predictive role to anti-HER3 therapies. Oncotarget. 2016;7:45042–51.
Ogier C, Colombo PE, Bousquet C, Canterel-Thouennon L, Sicard P, Garambois V, et al. Targeting the NRG1/HER3 pathway in tumor cells and cancer-associated fibroblasts with an anti-neuregulin 1 antibody inhibits tumor growth in pre-clinical models of pancreatic cancer. Cancer Lett. 2018;432:227–36.
Yun S, Koh J, Nam SK, Park JO, Lee SM, Lee K, et al. Clinical significance of overexpression of NRG1 and its receptors, HER3 and HER4, in gastric cancer patients. Gastric Cancer. 2018;21:225–36.
Wilson TR, Lee DY, Berry L, Shames DS, Settleman J. Neuregulin-1-mediated autocrine signaling underlies sensitivity to HER2 kinase inhibitors in a subset of human cancers. Cancer Cell. 2011;20:158–72.
Zhang K, Wong P, Zhang L, Jacobs B, Borden EC, Aster JC, et al. A Notch1-neuregulin1 autocrine signaling loop contributes to melanoma growth. Oncogene. 2012;31:4609–18.
Shee K, Yang W, Hinds JW, Hampsch RA, Varn FS, Traphagen NA, et al. Therapeutically targeting tumor microenvironment-mediated drug resistance in estrogen receptor-positive breast cancer. J Exp Med. 2018;215:895–910.
Chua YL, Ito Y, Pole JC, Newman S, Chin SF, Stein RC, et al. The NRG1 gene is frequently silenced by methylation in breast cancers and is a strong candidate for the 8p tumour suppressor gene. Oncogene. 2009;28:4041–52.
Pietras K, Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316:1324–31.
Orimo A, Weinberg RA. Heterogeneity of stromal fibroblasts in tumors. Cancer Biol Ther. 2007;6:618–9.
Cortez E, Roswall P, Pietras K. Functional subsets of mesenchymal cell types in the tumor microenvironment. Semin Cancer Biol. 2014;25:3–9.
Augsten M. Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front Oncol. 2014;4:62.
Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20:174–86.
Schneeweiss A, Park-Simon TW, Albanell J, Lassen U, Cortes J, Dieras V. et al. Phase Ib study evaluating safety and clinical activity of the anti-HER3 antibody lumretuzumab combined with the anti-HER2 antibody pertuzumab and paclitaxel in HER3-positive, HER2-low metastatic breast cancer. Invest new drugs. 2018;36:848–59.
Meulendijks D, Jacob W, Voest EE, Mau-Sorensen M, Martinez-Garcia M, Taus A, et al. Phase Ib study of lumretuzumab plus cetuximab or erlotinib in solid tumor patients and evaluation of HER3 and heregulin as potential biomarkers of clinical activity. Clin Cancer Res. 2017;23:5406–15.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52.
Cancer Genome, Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Casey T, Bond J, Tighe S, Hunter T, Lintault L, Patel O, et al. Molecular signatures suggest a major role for stromal cells in development of invasive breast cancer. Breast Cancer Res Treat. 2009;114:47–62.
Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7.
Vargas AC, McCart Reed AE, Waddell N, Lane A, Reid LE, Smart CE, et al. Gene expression profiling of tumour epithelial and stromal compartments during breast cancer progression. Breast Cancer Res Treat. 2012;135:153–65.
Liu H, Dowdle JA, Khurshid S, Sullivan NJ, Bertos N, Rambani K, et al. Discovery of stromal regulatory networks that suppress ras-sensitized epithelial cell proliferation. Dev Cell. 2017;41:392–407.e396.
Weinstein IB, Begemann M, Zhou P, Han EK, Sgambato A, Doki Y, et al. Disorders in cell circuitry associated with multistage carcinogenesis: exploitable targets for cancer prevention and therapy. Clin Cancer Res: Off J Am Assoc Cancer Res. 1997;3:2696–702.
Mirschberger C, Schiller CB, Schraml M, Dimoudis N, Friess T, Gerdes CA, et al. RG7116, a therapeutic antibody that binds the inactive HER3 receptor and is optimized for immune effector activation. Cancer Res. 2013;73:5183–94.
Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5:317–28.
Walker RA. The complexities of breast cancer desmoplasia. Breast Cancer Res. 2001;3:143–5.
Sonntag J, Schluter K, Bernhardt S, Korf U. Subtyping of breast cancer using reverse phase protein arrays. Expert Rev Proteom. 2014;11:757–70.
Kim J, Jeong H, Lee Y, Kim C, Kim H, Kim A. HRG-beta1-driven ErbB3 signaling induces epithelial-mesenchymal transition in breast cancer cells. BMC Cancer. 2013;13:383.
Koutsandreas TBI, Pilalis E, Valavanis I, Papadodima O, Chatziioannou A. Analyzing and visualizing genomic complexity for the derivation of the emergent molecular networks. Int J Monit Surveill Technol Res. 2016;4:30–49.
Loeb JA, Susanto ET, Fischbach GD. The neuregulin precursor proARIA is processed to ARIA after expression on the cell surface by a protein kinase C-enhanced mechanism. Mol Cell Neurosci. 1998;11:77–91.
Wernig G, Chen SY, Cui L, Van Neste C, Tsai JM, Kambham N, et al. Unifying mechanism for different fibrotic diseases. Proc Natl Acad Sci USA. 2017;114:4757–62.
Lisanti MP, Tsirigos A, Pavlides S, Reeves KJ, Peiris-Pages M, Chadwick AL, et al. JNK1 stress signaling is hyper-activated in high breast density and the tumor stroma: connecting fibrosis, inflammation, and stemness for cancer prevention. Cell Cycle. 2014;13:580–99.
Mishra R, Patel H, Alanazi S, Yuan L, Garrett JT. HER3 signaling and targeted therapy in cancer. Oncol Rev. 2018;12:355.
Karachaliou N, Lazzari C, Verlicchi A, Sosa AE, Rosell R. HER3 as a therapeutic target in cancer. BioDrugs. 2017;31:63–73.
Liu X, Liu S, Lyu H, Riker AI, Zhang Y, Liu B. Development of effective therapeutics targeting HER3 for cancer treatment. Biol Proced Online. 2019;21:5.
Sheng Q, Liu X, Fleming E, Yuan K, Piao H, Chen J, et al. An activated ErbB3/NRG1 autocrine loop supports in vivo proliferation in ovarian cancer cells. Cancer Cell. 2010;17:298–310.
Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.
Sadlonova A, Bowe DB, Novak Z, Mukherjee S, Duncan VE, Page GP, et al. Identification of molecular distinctions between normal breast-associated fibroblasts and breast cancer-associated fibroblasts. Cancer Microenviron. 2009;2:9–21.
Saadi A, Shannon NB, Lao-Sirieix P, O’Donovan M, Walker E, Clemons NJ, et al. Stromal genes discriminate preinvasive from invasive disease, predict outcome, and highlight inflammatory pathways in digestive cancers. Proc Natl Acad Sci USA. 2010;107:2177–82.
Berdiel-Acer M, Sanz-Pamplona R, Calon A, Cuadras D, Berenguer A, Sanjuan X, et al. Differences between CAFs and their paired NCF from adjacent colonic mucosa reveal functional heterogeneity of CAFs, providing prognostic information. Mol Oncol. 2014;8:1290–305.
Costea DE, Hills A, Osman AH, Thurlow J, Kalna G, Huang X, et al. Identification of two distinct carcinoma-associated fibroblast subtypes with differential tumor-promoting abilities in oral squamous cell carcinoma. Cancer Res. 2013;73:3888–901.
Patel AK, Vipparthi K, Thatikonda V, Arun I, Bhattacharjee S, Sharan R, et al. A subtype of cancer-associated fibroblasts with lower expression of alpha-smooth muscle actin suppresses stemness through BMP4 in oral carcinoma. Oncogenesis. 2018;7:78.
Morsing M, Klitgaard MC, Jafari A, Villadsen R, Kassem M, Petersen OW, et al. Evidence of two distinct functionally specialized fibroblast lineages in breast stroma. Breast Cancer Res. 2016;18:108.
Su S, Chen J, Yao H, Liu J, Yu S, Lao L, et al. CD10(+)GPR77(+) Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell. 2018;172:841–856.e816.
Parra LM, Hartmann M, Schubach S, Li Y, Herrlich P, Herrlich A. Distinct Intracellular Domain Substrate Modifications Selectively Regulate Ectodomain Cleavage of NRG1 or CD44. Mol Cell Biol. 2015;35:3381–95.
Karagiannis GS, Poutahidis T, Erdman SE, Kirsch R, Riddell RH, Diamandis EP. Cancer-associated fibroblasts drive the progression of metastasis through both paracrine and mechanical pressure on cancer tissue. Mol Cancer Res. 2012;10:1403–18.
Kim YH, Lee SB, Shim S, Kim A, Park JH, Jang WS, et al. Hyaluronic acid synthase 2 promotes malignant phenotypes of colorectal cancer cells through transforming growth factor beta signaling. Cancer Sci. 2019;110:2226–36.
Zhang Z, Tao D, Zhang P, Liu X, Zhang Y, Cheng J, et al. Hyaluronan synthase 2 expressed by cancer-associated fibroblasts promotes oral cancer invasion. J Exp Clin Cancer Res. 2016;35:181.
Mei L, Nave KA. Neuregulin-ERBB signaling in the nervous system and neuropsychiatric diseases. Neuron. 2014;83:27–49.
Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–52.
Ledonne A, Mercuri NB. On the modulatory roles of neuregulins/ErbB signaling on synaptic plasticity. Int J Mol Sci. 2019;21:275.
Kudo D, Suto A, Hakamada K. The development of a novel therapeutic strategy to target hyaluronan in the extracellular matrix of pancreatic ductal adenocarcinoma. Int J Mol Sci. 2017;18:600.
Yoshida E, Kudo D, Nagase H, Suto A, Shimoda H, Suto S, et al. 4-Methylumbelliferone decreases the hyaluronan-rich extracellular matrix and increases the effectiveness of 5-fluorouracil. Anticancer Res. 2018;38:5799–804.
Sahin O, Lobke C, Korf U, Appelhans H, Sultmann H, Poustka A, et al. Combinatorial RNAi for quantitative protein network analysis. Proc Natl Acad Sci USA. 2007;104:6579–84.
Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–261.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29.
Acknowledgements
We thank N.Erdem and C.Koerner for valuable discussions, the Department of Gynecology, Martin-Luther-University Halle-Wittenberg, Halle (Saale), Germany for providing tumour tissue and clinical data, Breast Cancer Now Tissue Bank (https://breastcancernow.org/) for providing primary fibroblasts. E.Reinz and S.Kemmer for contributing to Fig. S1C. D.Heiss and S.Karolus provided excellent technical assisstance.The Genomics & Proteomics as well as the Light Microscopy Core Facilities at German Cancer Research Center performed excellent services. This work was supported by the German Federal Ministry of Education and Research (e:Med FKZ: 031A429).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Berdiel-Acer, M., Maia, A., Hristova, Z. et al. Stromal NRG1 in luminal breast cancer defines pro-fibrotic and migratory cancer-associated fibroblasts. Oncogene 40, 2651–2666 (2021). https://doi.org/10.1038/s41388-021-01719-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01719-3