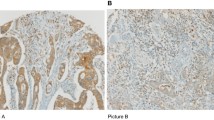
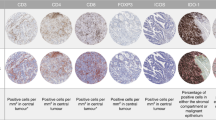
Abstract
Colorectal cancer (CRC) is characterized by a heterogeneous tumor microenvironment (TME) that regulates cancer progression and therapeutic response. Overexpression of FOXP3 and CTLA4 is associated with immunosuppressive TME and poor prognosis in many cancer types. However, opposite results were reported in CRC. Thus, we performed comprehensive analyses to evaluate the exact prognostic value of FOXP3 and CTLA4 in CRC. Here, the expression levels of FOXP3 and CTLA4 were used to construct a subtyping system based on >1200 CRC patients from multiple independent public datasets. We revealed that, in CRC patients with relatively high expression of FOXP3, there exist two different subpopulations with opposite survival patterns according to CLTA4 expression. We further established a method for evaluating all cohorts and identified a novel FOXP3HighCTLA4High* CRC risk subpopulation that accounts for 5–10% of CRC patients. Moreover, different methods of functional enrichment and immune evaluation were used to analyze the TME characteristics of different FOXP3/CTLA4 subtypes. The FOXP3HighCTLA4High* CRC risk subpopulation was characterized by an immune overdrive TME phenotype, including high immune cell infiltration, low tumor purity, high immune checkpoint levels, and TGF-β activation. Finally, the constructed FOXP3/CTLA4 subtyping system was further validated by quantitative RT-PCR, immunochemistry staining, and multicolor immunofluorescence in an independent CRC cohort we collected. This high-risk subpopulation was also observed in kidney cancers and low-grade glioma patients by a Pan-cancer analysis. Together, our study revealed that the established FOXP3/CTLA4 molecular subtyping system could be used to select treatment and management strategies for CRC and other cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235–46.
Giraldo NA, Sanchez-Salas R, Peske JD, Vano Y, Becht E, Petitprez F, et al. The clinical role of the TME in solid cancer. Br J Cancer. 2019;120:45–53.
Yin Y, Yao S, Hu Y, Feng Y, Li M, Bian Z, et al. The immune-microenvironment confers chemoresistance of colorectal cancer through macrophage-derived IL6. Clin Cancer Res. 2017;23:7375–87.
Riera-Domingo C, Audigé A, Granja S, Cheng W-C, Ho P-C, Baltazar F, et al. Immunity, hypoxia, and metabolism-the ménage à trois of cancer: implications for immunotherapy. Physiol Rev. 2020;100:1–102.
Becht E, de Reynies A, Giraldo NA, Pilati C, Buttard B, Lacroix L, et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res. 2016;22:4057–66.
Iglesia MD, Parker JS, Hoadley KA, Serody JS, Perou CM, Vincent BG. Genomic analysis of immune cell infiltrates across 11 tumor types. J Natl Cancer Inst. 2016;108:djw144.
Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–62.
Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44:698–711.
Pages F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–39.
Toor SM, Sasidharan Nair V, Decock J, Elkord E. Immune checkpoints in the tumor microenvironment. Semin Cancer Biol. 2020;65:1–12.
Deng L, Gyorffy B, Na F, Chen B, Lan J, Xue J, et al. Association of PDCD1 and CTLA-4 Gene expression with clinicopathological factors and survival in non¨CSmall-Cell lung cancer: results from a large and pooled microarray database. J Thorac Oncol. 2015;10:1020–6.
Heinzerling L, Ott PA, Hodi FS, Husain AN, Tajmir-Riahi A, Tawbi H, et al. Cardiotoxicity associated with CTLA4 and PD1 blocking immunotherapy. J Immunother Cancer. 2016;4:50.
Herrmann A, Lahtz C, Nagao T, Song JY, Chan WC, Lee H, et al. CTLA4 promotes Tyk2-STAT3¨C dependent B-cell oncogenicity. Cancer Res. 2017;77:5118–28.
Yu H, Yang J, Jiao S, Li Y, Zhang W, Wang J. Cytotoxic T lymphocyte antigen 4 expression in human breast cancer: implications for prognosis. Cancer Immunol Immunother. 2015;64:853–60.
Lu L, Bai Y, Wang Z. Elevated T cell activation score is associated with improved survival of breast cancer. Breast Cancer Res Treat. 2017;164:689–96.
Xiang Z, Chen W, Zhang J, Song S, Xia G-K, Huang X-Y, et al. Identification of discrepancy between CTLA4 expression and CTLA4 activation in gastric cancer. Immunopharmacol Immunotoxicol. 2018;41:386–93.
Zaravinos A, Roufas C, Nagara M, de Lucas Moreno B, Oblovatskaya M, Efstathiades C, et al. Cytolytic activity correlates with the mutational burden and deregulated expression of immune checkpoints in colorectal cancer. J Exp Clin Cancer Res. 2019;38:364.
Ciardiello D, Vitiello PP, Cardone C, Martini G, Troiani T, Martinelli E, et al. Immunotherapy of colorectal cancer: challenges for therapeutic efficacy. Cancer Treat Rev. 2019;76:22–32.
Raffin C, Vo LT, Bluestone JA. Treg cell-based therapies: challenges and perspectives. Nat Rev Immunol. 2020;20:158–72.
Punt S, Houwing-Duistermaat JJ, Schulkens IA, Thijssen VL, Osse EM, de Kroon CD, et al. Correlations between immune response and vascularization qRT-PCR gene expression clusters in squamous cervical cancer. Mol Cancer. 2015;14:71.
Wei YF, Chu CY, Chang CC, Lin SH, Su WC, Tseng YL, et al. Different pattern of PD-L1, IDO, and FOXP3 Tregs expression with survival in thymoma and thymic carcinoma. Lung Cancer. 2018;125:35–42.
Wolf D. The expression of the regulatory T cell-specific forkhead box transcription factor Foxp3 is associated with poor prognosis in ovarian cancer. Clin Cancer Res. 2005;11:8326–31.
Shi J-Y, Ma L-J, Zhang J-W, Duan M, Ding Z-B, Yang L-X, et al. FOXP3 is a HCC suppressor gene and acts through regulating the TGF-β/Smad2/3 signaling pathway. BMC Cancer. 2017;17:648.
Yang S, Liu Y, Li M-Y, Ng CSH, Yang S-l, Wang S, et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer. 2017;16:124.
Lal A, Chan L, Devries S, Chin K, Scott GK, Benz CC, et al. FOXP3-positive regulatory T lymphocytes and epithelial FOXP3 expression in synchronous normal, ductal carcinoma in situ, and invasive cancer of the breast. Breast Cancer Res Treat. 2013;139:381–90.
Weller P, Bankfalvi A, Gu X, Dominas N, Lehnerdt GTF, Zeidler R, et al. The role of tumour FoxP3 as prognostic marker in different subtypes of head and neck cancer. Eur J Cancer. 2014;50:1291–300.
Ganapathi SK, Beggs AD, Hodgson SV, Kumar D. Expression and DNA methylation of TNF, IFNG and FOXP3 in colorectal cancer and their prognostic significance. Br J Cancer. 2014;111:1581–9.
Sun X, Feng Z, Wang Y, Qu Y, Gai Y. Expression of Foxp3 and its prognostic significance in colorectal cancer. Int J Immunopathol Pharm. 2017;30:201–6.
Chew A, Salama P, Robbshaw A, Klopcic B, Zeps N, Platell C, et al. SPARC, FOXP3, CD8 and CD45 correlation with disease recurrence and long-term disease-free survival in colorectal cancer. PLoS ONE. 2011;6:e22047.
Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–82.
Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–10.
Petitprez F, Levy S, Sun CM, Meylan M, Linhard C, Becht E, et al. The murine microenvironment cell population counter method to estimate abundance of tissue-infiltrating immune and stromal cell populations in murine samples using gene expression. Genome Med. 2020;12:86.
Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6.
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou YTH, et al. The immune landscape of cancer. Immunity .2018;48:812–30.e814.
Eide PW, Bruun J, Lothe RA, Sveen A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci Rep. 2017;7:16618.
Batlle E, Massague J. Transforming growth factor-beta signaling in immunity and cancer. Immunity. 2019;50:924–40.
Lee HO, Hong Y, Etlioglu HE, Cho YB, Pomella V, Van den Bosch B, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet. 2020;52:594–603.
Wu C, Li M, Meng H, Liu Y, Niu W, Zhou Y, et al. Analysis of status and countermeasures of cancer incidence and mortality in China. Sci China Life Sci. 2019;62:640–7.
Freeman ZT, Nirschl TR, Hovelson DH, Johnston RJ, Engelhardt JJ, Selby MJ, et al. A conserved intratumoral regulatory T cell signature identifies 4-1BB as a pan-cancer target. J Clin Invest. 2020;130:1405–16.
Ramos-Casals M, Brahmer JR, Callahan MK, Flores-Chavez A, Keegan N, Khamashta MA, et al. Immune-related adverse events of checkpoint inhibitors. Nat Rev Dis Prim. 2020;6:38.
Andrews LP, Yano H, Vignali DAA. Inhibitory receptors and ligands beyond PD-1, PD-L1 and CTLA-4: breakthroughs or backups. Nat Immunol. 2019;20:1425–34.
Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17:79–92.
Fakih M, Ouyang C, Wang C, Tu TY, Gozo MC, Cho M, et al. Immune overdrive signature in colorectal tumor subset predicts poor clinical outcome. J Clin Invest. 2019;129:4464–76.
Melaiu O, Lucarini V, Giovannoni R, Fruci D, Gemignani F. News on immune checkpoint inhibitors as immunotherapy strategies in adult and pediatric solid tumors. Semin Cancer Biol. 2020.
Bai X, Yi M, Jiao Y, Chu Q, Wu K. Blocking TGF-beta signaling to enhance the efficacy of immune checkpoint inhibitor. Onco Targets Ther. 2019;12:9527–38.
Fiegle E, Doleschel D, Koletnik S, Rix A, Weiskirchen R, Borkham-Kamphorst E, et al. Dual CTLA-4 and PD-L1 blockade inhibits tumor growth and liver metastasis in a highly aggressive orthotopic mouse model of colon cancer. Neoplasia .2019;21:932–44.
Rotte A. Combination of CTLA-4 and PD-1 blockers for treatment of cancer. J Exp Clin Cancer Res. 2019;38:255.
Xie YH, Chen YX, Fang JY. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther. 2020;5:22.
Wu F, Wang ZL, Wang KY, Li GZ, Chai RC, Liu YQ, et al. Classification of diffuse lower-grade glioma based on immunological profiling. Mol Oncol. 2020;14:2081–95.
Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell .2018;173:400–16. e411.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. Biol. 2012;16:284–7.
Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinforma. 2013;14:7.
Yin Y, Zhang B, Wang W, Fei B, Quan C, Zhang J, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res. 2014;20:6187–99.
Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70:46–58.
Cui K, Liu C, Li X, Zhang Q, Li Y. Comprehensive characterization of the rRNA metabolism-related genes in human cancer. Oncogene. 2020;39:786–800.
Acknowledgements
We thank the TCGA, GEO, and CPTAC project. We thank the Affiliated Hospital of Jiangnan University for providing the CRC samples. This work was supported by grants from the National Natural Science Foundation of China (82002550, 81672328, and 81972220), Jiangsu Key Research and Development Plan (BE2019632), Medical Key Professionals Program of Jiangsu Province (AF052141), National First-class Discipline Program of Food Science and Technology (JUFSTR20180101), and Wuxi Medical Innovation Team (CXTP003).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Cui, K., Yao, S., Zhang, H. et al. Identification of an immune overdrive high-risk subpopulation with aberrant expression of FOXP3 and CTLA4 in colorectal cancer. Oncogene 40, 2130–2145 (2021). https://doi.org/10.1038/s41388-021-01677-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01677-w
This article is cited by
-
Palmitoylation landscapes across human cancers reveal a role of palmitoylation in tumorigenesis
Journal of Translational Medicine (2023)
-
The heterogeneity of tumour immune microenvironment revealing the CRABP2/CD69 signature discriminates distinct clinical outcomes in breast cancer
British Journal of Cancer (2023)
-
EXOSC8 promotes colorectal cancer tumorigenesis via regulating ribosome biogenesis-related processes
Oncogene (2022)
-
A novel high-risk subpopulation identified by CTSL and ZBTB7B in gastric cancer
British Journal of Cancer (2022)