Abstract
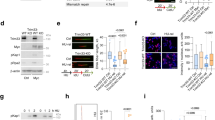
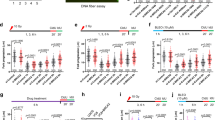
The Mdm4 (alias MdmX) oncoprotein, like its paralogue and interaction partner Mdm2, antagonizes the tumor suppressor p53. p53-independent roles of the Mdm proteins are emerging, and we have reported the ability of Mdm2 to modify chromatin and to support DNA replication by suppressing the formation of R-loops (DNA/RNA-hybrids). We show here that the depletion of Mdm4 in p53-deficient cells compromises DNA replication fork progression as well. Among various deletion mutants, only full-length Mdm4 was able to support DNA replication fork progression. Co-depletion of Mdm4 and Mdm2 further impaired DNA replication, and the overexpression of each partially compensated for the other’s loss. Despite impairing replication, Mdm4 depletion only marginally hindered cell proliferation, likely due to compensation through increased firing of replication origins. However, depleting Mdm4 sensitized p53−/− cells to the nucleoside analog gemcitabine, raising the future perspective of using Mdm4 inhibitors as chemosensitizers. Mechanistically, Mdm4 interacts with members of the Polycomb Repressor Complexes and supports the ubiquitination of H2A, thereby preventing the accumulation of DNA/RNA-hybrids. Thus, in analogy to previously reported activities of Mdm2, Mdm4 enables unperturbed DNA replication through the avoidance of R-loops.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci. 2003;100:12009–14.
Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–43.
Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–6.
Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med. 2012;18:1239–47.
Han X, Medeiros LJ, Zhang YH, You MJ, Andreeff M, Konopleva M, et al. High expression of human homologue of murine double minute 4 and the short splicing variant, hdm4-s, in bone marrow in patients with acute myeloid leukemia or myelodysplastic syndrome. Clin Lymphoma Myeloma Leuk. 2016;16:S30–8.
Matijasevic Z, Krzywicka-Racka A, Sluder G, Jones SN. MdmX regulates transformation and chromosomal stability in p53-deficient cells. Cell Cycle. 2008;7:2967–73.
Carillo Alexia M. Bouska Alzssa, Arrate Maria Pia ECM. NIH Public Access. Oncogene 2015;34:846–56.
Strachan GD, Jordan-Sciutto KL, Rallapalli R, Tuan RS, Hall DJ. The E2F-1 transcription factor is negatively regulated by its interaction with the MDMX protein. J Cell Biochem. 2003;88:557–68.
Zhang H, Hu L, Qiu W, Deng T, Zhang Y, Bergholz J, et al. MDMX exerts its oncogenic activity via suppression of retinoblastoma protein. Oncogene. 2015;34:5560–9.
Jin Y, Zeng SX, Sun X-X, Lee H, Blattner C, Xiao Z, et al. MDMX promotes proteasomal turnover of p21 at G1 and early S phases independently of, but in cooperation with, MDM2. Mol Cell Biol. 2008;28:1218–29.
Bohlman S, Manfredi JJ. p53-independent effects of Mdm2. In: Deb S, Deb S (eds). Mutant p53 and MDM2 in Cancer. Subcellular Biochemistry, vol 85. (Springer, Dordrecht, 2014) pp 235–46.
Alt JR, Bouska A, Fernandez MR, Cerny RL, Xiao H, Eischen CM. Mdm2 Binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J Biol Chem. 2005;280:18771–81.
Bouska A, Lushnikova T, Plaza S, Eischen CM. Mdm2 promotes genetic instability and transformation independent of p53. Mol Cell Biol. 2008;28:4862–74.
Klusmann I, Rodewald S, Müller L, Friedrich M, Wienken M, Li Y, et al. p53 activity results in DNA replication fork processivity. Cell Rep. 2016;17:1845–57.
Wienken M, Dickmanns A, Nemajerova A, Kramer D, Najafova Z, Weiss M, et al. MDM2 associates with polycomb repressor complex 2 and enhances stemness-promoting chromatin modifications independent of p53. Mol Cell 2016;61:68–83.
Klusmann I, Wohlberedt K, Magerhans A, Teloni F, Korbel JO, Altmeyer M, et al. Chromatin modifiers Mdm2 and RNF2 prevent RNA:DNA hybrids that impair DNA replication. Proc Natl Acad Sci. 2018;115:E11311–20.
Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011;25:2041–56.
Shvarts A, Bazuine M, Dekker P, Ramos YFM, Steegenga WT, Merckx G, et al. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics 1997;43:34–42.
Gazdar AF, Gao B, Minna JD. Lung cancer cell lines: useless artifacts or invaluable tools for medical science? Lung Cancer 2010;68:309–18.
Gradiz R, Silva HC, Carvalho L, Botelho MF, Mota-Pinto A. MIA PaCa-2 and PANC-1 – pancreas ductal adenocarcinoma cell lines with neuroendocrine differentiation and somatostatin receptors. Sci Rep. 2016;6:21648.
Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 2001;29:92–5.
Meulmeester E, Frenk R, Stad R, de Graaf P, Marine J-C, Vousden KH, et al. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol Cell Biol. 2003;23:4929–38.
Dobbelstein M, Sørensen CS. Exploiting replicative stress to treat cancer. Nat Rev Drug Discov. 2015;14:405–23.
Blow JJ, Ge XQ, Jackson DA. How dormant origins promote complete genome replication. Trends Biochem Sci. 2011;36:405–14.
Quinet A, Carvajal-Maldonado D, Lemacon D, Vindigni A. DNA fiber analysis: mind the gap! In: Eichman BF (ed). DNA Repair Enzymes: Cell, Molecular, and Chemical Biology. Methods in Enzymology, vol 591. (Elsevier, Amsterdam, 2017) pp 55–82.
Plunkett W, Huang P, Searcy CE, Gandhi V. Gemcitabine: preclinical pharmacology and mechanisms of action. Semin Oncol 1996;23:3–15.
Wen W, Peng C, Kim MO, Ho Jeong C, Zhu F, Yao K, et al. Knockdown of RNF2 induces apoptosis by regulating MDM2 and p53 stability. Oncogene. 2014;33:421–8.
Kuser-Abali G, Gong L, Yan J, Liu Q, Zeng W, Williamson A, et al. An EZH2-mediated epigenetic mechanism behind p53-dependent tissue sensitivity to DNA damage. Proc Natl Acad Sci. 2018;115:201719532.
Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24.
Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, et al. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods. 1986;89:123–30.
El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–58.
Fischer M, Steiner L, Engeland K. The transcription factor p53: not a repressor, solely an activator. Cell Cycle. 2014;13:3037–58.
Momand J, Villegas A, Belyi VA. The evolution of MDM2 family genes. Gene. 2011;486:23–30.
Fåhraeus R, Olivares-Illana V. MDM2’s social network. Oncogene. 2014;33:4365–76.
Riley MF, Lozano G. The many faces of MDM2 binding partners. Genes Cancer. 2012;3:226–39.
Haupt S, Mejía-Hernández JO, Vijayakumaran R, Keam SP, Haupt Y. The long and the short of it: the MDM4 tail so far. J Mol Cell Biol. 2019;11:231–44.
Stad R, Little NA, Xirodimas DP, Frenk R, Eb AJ, van der, Lane DP, et al. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep. 2001;2:1029.
Lee J, Dunphy WG. The Mre11-Rad50-Nbs1 (MRN) complex has a specific role in the activation of Chk1 in response to stalled replication forks. Mol Biol Cell. 2013;24:1343–53.
Dewaele M, Tabaglio T, Willekens K, Bezzi M, Teo SX, Low DHP, et al. Antisense oligonucleotide–mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest. 2015;126:68–84.
Matijasevic Z, Steinman HA, Hoover K, Jones SN. MdmX promotes bipolar mitosis to suppress transformation and tumorigenesis in p53-deficient cells and mice. Mol Cell Biol. 2008;28:1265–73.
Marine J-C, Jochemsen AG. MDMX (MDM4), a Promising target for p53 reactivation therapy and beyond. Cold Spring Harb Perspect Med. 2016;6. https://doi.org/10.1101/cshperspect.a026237.
Valentin-Vega YA, Box N, Terzian T, Lozano G. Mdm4 loss in the intestinal epithelium leads to compartmentalized cell death but no tissue abnormalities. Differentiation. 2009;77:442–9.
Garcia D, Warr MR, Martins CP, Brown Swigart L, Passegué E, Evan GI. Validation of MdmX as a therapeutic target for reactivating p53 in tumors. Genes Dev. 2011;25:1746–57.
Heijkants RC, Nieveen M, Hart KC’t, Teunisse AFAS, Jochemsen AG. Targeting MDMX and PKCδ to improve current uveal melanoma therapeutic strategies. Oncogenesis. 2018;7:33.
Miranda PJ, Buckley D, Raghu D, Pang J-MB, Takano EA, Vijayakumaran R, et al. MDM4 is a rational target for treating breast cancers with mutant p53. J Pathol. 2017;241:661–70.
Park DE, Cheng J, Berrios C, Montero J, Cortés-Cros M, Ferretti S, et al. Dual inhibition of MDM2 and MDM4 in virus-positive Merkel cell carcinoma enhances the p53 response. Proc Natl Acad Sci. 2019;116:1027–32.
Carvajal LA, Neriah DBen, Senecal A, Benard L, Thiruthuvanathan V, Yatsenko T, et al. Dual inhibition of MDMX and MDM2 as a therapeutic strategy in leukemia. Sci Transl Med. 2018;10:eaao3003.
Pellegrino M, Mancini F, Lucà R, Coletti A, Giacchè N, Manni I, et al. Targeting the MDM2/MDM4 interaction interface as a promising approach for p53 reactivation therapy. Cancer Res. 2015;75:4560–72.
Bernal F, Wade M, Godes M, Davis TN, Whitehead DG, Kung AL, et al. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18:411–22.
Li Y, Yang J, Aguilar A, McEachern D, Przybranowski S, Liu L, et al. Discovery of MD-224 as a first-in-class, highly potent, and efficacious proteolysis targeting chimera murine double minute 2 degrader capable of achieving complete and durable tumor regression. J Med Chem. 2019;62:448–66.
Técher H, Koundrioukoff S, Azar D, Wilhelm T, Carignon S, Brison O, et al. Replication dynamics: Biases and robustness of DNA fiber analysis. J Mol Biol. 2013;425:4845–55.
Acknowledgements
We thank Guillermina Lozano for the MEFs with p53/Mdm4/Mdm2 deletions. pCMV-Flag-Mdm4 was a gift from Zhi-Min Yuan. pCMV-MDM2 was a gift from Bert Vogelstein (Addgene plasmid #16441), pCMV-MDM2(C464A) was provided by Tyler Jacks (Addgene plasmid #12086), pICE-RNaseHI-WT-NLS-mCherry (Addgene plasmid #60365) as well as pICE-RNaseHI-D10R-E48R-NLS-mCherry (Addgene plasmid #60367) were obtained from Patrick Calsou. H2A and EZH2 expression plasmids were from Titia Sixma (Addgene plasmids #63561 and #63564) and Kristian Helin (Addgene plasmid #24230), respectively. pLenti6/V5-DEST-RNF2 was a gift from Lynda Chin (Addgene plasmid #31216). This work was supported by the Deutsche Krebshilfe (to MD and KW), the Wilhelm Sander Stiftung, the Else Kröner Fresenius Stiftung, the Deutsche José Carreras Leukämie Stiftung, the Deutsche Forschungsgemeinschaft, the Boehringer Ingelheim Fonds (to IK) and the German Academic Scholarship Foundation (to KW). IK, PD, and VM were members of the IMPRS/MSc/PhD program Molecular Biology and IK, VM, CG and JC also of the Göttingen Graduate School GGNB Göttingen.
Author information
Authors and Affiliations
Contributions
KW, IK, and MD designed research; KW, IK, PD, KH, JC, AM, VM, and CG performed research; CME contributed expression constructs for Mdm4 mutants and DNA replication expertise; AGJ performed immunoprecipitation; KW and IK analyzed data; KW, IK, and MD wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wohlberedt, K., Klusmann, I., Derevyanko, P.K. et al. Mdm4 supports DNA replication in a p53-independent fashion. Oncogene 39, 4828–4843 (2020). https://doi.org/10.1038/s41388-020-1325-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-1325-1
This article is cited by
-
MDM4 was associated with poor prognosis and tumor-immune infiltration of cancers
European Journal of Medical Research (2024)
-
MDMX elevation by a novel Mdmx–p53 interaction inhibitor mitigates neuronal damage after ischemic stroke
Scientific Reports (2022)
-
The integrated stress response induces R-loops and hinders replication fork progression
Cell Death & Disease (2020)