Abstract
Kidney cancer incidences are rising globally, thereby fueling the demand for targeted therapies and precision medicine. In our previous work, we have identified and characterized the Ras-Association Domain Family encoding ten members that are often aberrantly expressed in human cancers. In this study, we created and analyzed the Rassf10 knockout mice. Here we show that Rassf10 haploinsufficiency promotes neoplasia formation in two established mouse cancer models (Rassf1A−/− and p53−/−). Haploinsufficient Rassf10 knockout mice were significantly prone to various diseases including lymphoma (Rassf1A−/− background) and thymoma (p53−/− background). Especially Rassf10−/− and p53-deficient mice exhibited threefold increased rates of kidney cysts compared with p53−/− controls. Moreover, we observed that in human kidney cancer, RASSF10 is frequently epigenetically inactivated by its CpG island promoter hypermethylation. Primary tumors of renal clear cell and papillary cell carcinoma confirmed that RASSF10 methylation is associated with decreased expression in comparison to normal kidney tissue. In independent data sets, we could validate that RASSF10 inactivation clinically correlated with decreased survival and with progressed disease state of kidney cancer patients and polycystic kidney size. Functionally, we revealed that the loss of Rassf10 was significantly associated with upregulation of KRAS signaling and MYC expression. In summary, we could show that Rassf10 functions as a haploinsufficient tumor suppressor. In combination with other markers, RASSF10 silencing can serve as diagnostic and prognostic cancer biomarker in kidney diseases.
Similar content being viewed by others
Introduction
Cancer still is a leading cause of morbidity and mortality worldwide. Fourteen million new cases per year, more than 8 million deaths, an economic impact above US$1 trillion and numbers are expected to rise further in the next years [1]. Expectedly, there has been a great scientific interest in the underlying genetic changes of cancer development, in the hope to decelerate these rising cancer numbers by targeted approaches. Biomarkers are of use at all disease stages and have an impact on most cancer patients’ diagnosis/treatment nowadays [2, 3]. Biomarkers like BRCA and HER-2 in breast cancer [4], PSA in prostate cancer [5] and EGFR in lung cancer [6] are already being valued in the clinic. Our group focuses on epigenetically inactivated tumor suppressors as candidate biomarkers [7,8,9,10,11,12,13], which are predicted to play a prominent role in the near future [14]. The tumor-suppressor RASSF10 is a member of the tumor-suppressor family Ras-Association Domain Family (RASSF) [15, 16]. The family has gained attention since its first description in the year 2000 [7]. The RASSFs differ substantially in their tumor-suppressor pathways [15,16,17]. RASSF10, contains its RA-domain N-terminally and lacks catalytically active domains [15, 16]. RASSF10, located at 11p15.3, contains a large CpG island promoter >2 kb (NCBI, Entrez Gene; Supplementary Fig. S1). Epigenetic inactivation of RASSF10 by its promoter hypermethylation is a frequent event in pathogenesis of human cancers [18,19,20,21,22,23]. In previous studies, we have shown that the RASSF10 promoter is methylated in patient tumors samples of the adrenal gland [24], head and neck [20], sarcoma [20], pancreas carcinoma [20], and Merkel cell carcinoma [25]. We showed the epigenetic inactivation of RASSF10 in breast cancer [26], lung cancer [20], skin cancer [27], and thyroid cancer [21] and showed that RASSF10 inhibited the growth of breast cancer [26], pancreas carcinoma, and sarcoma cell lines [20]. So far, RASSF10 has only been studied in vitro and inconsistent pathways of action were suggested like cAMP-PKA signaling [20], MMP2 [28], p53 [29], and JNK [30]. In vivo analyses of RASSF10 were missing and the function of RASSF10 has not been analyzed in kidney cancer.
In our study, we generated the first Rassf10 knockout mouse model/animal model and we present Rassf10 as a novel haploinsufficient tumor suppressor in vivo. The loss of Rassf10 is linked to upregulation of KRAS signaling and MYC induction. Moreover, we revealed that RASSF10 is frequently epigenetically inactivated in kidney cancer, and we show the clinical potential of RASSF10 as a biomarker in different kidney diseases.
Results
Generation and characterization of the Rassf10 knockout
To analyze the tumor-suppressor function of RASSF10 in an animal model, we generated the Rassf10 knockout mouse. RASSF10 shares 86% amino acid identity between humans and mice regarding its sequence, domain structure (RA domain 87%) and the same genomic arrangement with a large CpG island covering its promoter (NCBI, UCSC genome browser, Supplementary Fig. S1). Rassf10 is broadly expressed in adult wildtype mouse tissues (strain C57BL/6) and predominantly in lung, kidney, and thymus (Fig. 1a). During embryonal stages E15.5/E18.5 and newborn, we found that Rassf10 is expressed in lung, kidney, and the developing skin (Fig. 1b). These expression data were verified by RNA in situ hybridization for Rassf10 in embryos (E9.5/E10.5/E11.5; Supplementary Fig. 1C). According sections detected Rassf10 in the developing kidney (mesonephros) and the lung bud/trachea. Due to Rassf10’s expression pattern, we generated a constitutive Rassf10 knockout (Supplementary Fig. S2). We interbred Rassf10+/− animals (Fig. 1) and observed an alteration in the Mendelian ratio of the offspring (Table 1). The number of born Rassf10−/− animals was significantly diminished (p < 0.04 χ2; Rassf10−/− observed n = 67 vs. expected n = 87). We did not observe maldeveloped embryos (E14), implicating that Rassf10 loss impairs earlier embryogenesis. The surviving Rassf10−/− animals developed normally (Supplementary Fig. S3) are fertile and showed no obvious phenotype.
a RNA expression of Rassf10 across mouse tissues from two adult wt C57BL/6 mice as normalized to Gapdh level (lung = 1). b Rassf10 is expressed in the developing mouse shown for two embryonal stages (E15.5 and E18.5) and in newborn (P0). Rassf10-RNA levels are normalized to Gapdh expression. c Rassf10 is expressed in whole mouse embryos by RNA in situ hybridization at E9.5, E10.5, and E11.5 (2 mm white bar) and according section is shown (1 mm black bar).
The Rassf10 knockout promotes neoplasia in cancer prone mice
We tested if Rassf10 loss induces spontaneous tumor formation in aging mice (Fig. 2a). At 80 weeks animals showed no differences between wt (n = 27) and Rassf10−/− (n = 44), regarding the occurrence of various diseases/neoplasia (diseased wt 56% vs. Rassf10−/− 57%). Therefore, we decided on a double knockout approach of tumor suppressors to accelerate disease/tumor onset, consistent with the cancer hypothesis of progressive accumulation of mutations [31]. We chose the p53−/− (tumors <6 months [32, 33]) and separate Rassf1A−/− (R1A; tumors at 18–20 months [34]) background to cross into our Rassf10 knockout (Fig. 2a). After 20 weeks for the p53−/− double knockout and 80 weeks for the Rassf1A−/− double knockout, all animals were sacrificed and full necropsy was performed (Table 2). In the Rassf1A−/− tumor-suppressor background, the additional Rassf10 knockout reduced significantly the overall survival (p = 0.018; Fig. 2b). We observed significantly diseased animals Rassf10+/− at 62% and Rassf10−/− at 59% vs. wt at 24% (Table 2). In detail Rassf10 knockout animals mostly suffered from lymphoma, and the mean size of measurable lymph nodes increased from 86 mg for Rassf10 wt to 285 mg Rassf10+/− to 401 mg for Rassf10−/− (Fig. 2c, d). The median lymph node weight was 53, 201, and 136 mg, respectively. Lymph nodes were significantly enlarged in 53% Rassf10+/− and 50% Rassf10−/− vs. 10% Rassf10+/+ (>100 mg; Table 2). Rassf10 loss also enlarged the spleen and was present in 53% Rassf10+/− and 40% Rassf10−/− vs. 28% Rassf10+/+ (Table 2 and Supplementary Fig. S4; >95 mg). The mean spleen weight was 126 mg for Rassf10+/+ and increased by Rassf10 loss for Rassf10+/− to 163 mg (Supplementary Fig. S4). The presence of lymphoma correlated with the enlargement of the spleen (splenomegaly; Supplementary Fig. S4B). In addition, Rassf1A knockout animals not only developed megaesophagus (reported earlier [35]), but also megaileum/megacolon at a total incidence 16%. This malformation of the digestive tract was Rassf10 independent and is a novel finding.
a Schematic representation of Rassf10 knockout mouse generation and experimental setup. Rassf10 knockout (KO) animals were subjected to aging, additional Rassf1A (R1A) knockout or p53 knockout. b Kaplan–Meier survival curves for Rassf10 KO mice reveal reduced survival vs. wt-Rassf10 animals in R1A−/− background (p = 0.018, log rank test). Number of animals are 12 Rassf10+/+, 71 Rassf10+/−/Rassf10−/−. The Y-axis starting at 50%. c Rassf10 KO mice suffered from increased size of lymph nodes in comparison to wildtype Rassf10 mice in R1A−/− background. Number of animals are 8 Rassf10+/+, 16 Rassf10+/−, and 24 Rassf10−/−. d The Rassf10 KO induces enlarged lymph nodes (lymphoma). Tissues were isolated during necropsy, fixed with formaldehyde, paraffin embedded, 10 µm sections were prepared and H&E stained before microscopic analysis.
The Rassf10 knockout promotes kidney neoplasia in p53-deficient mice
In the p53−/− tumor-suppressor background, survival analysis by Kaplan–Meier revealed that Rassf10 knockout animals (n = 77; Rassf10+/−/Rassf10−/−) had a reduced survival rate vs. Rassf10+/+ animals (n = 24, Fig. 3a, Table 2). However, this trend was not significant. We observed diseased animals at the following rates (p53−/− background): wt 58% (14/24), Rassf10+/− 81% (30/37), and Rassf10−/− 70% (28/40). In detail, we found that the thymus and the kidney were mostly affected by Rassf10 knockout (Fig. 3 and Table 2). Interestingly, enlarged thymus and thymoma were found in 24% Rassf10−/− (8/33), in 46% Rassf10+/− (12/26) but only in 11% Rassf10-wt (2/18) animals, significant regarding Rassf10+/− vs. wt (Table 2; Fig. 3b, c). Enlarged thymus were determined by a > 2.5x weight increase above average wt thymus (47.5 ± 2.4 mg Rassf10+/+; corrected by two thymoma; Supplementary Fig. S5). Rassf10 heterozygous knockout animals exhibited a dramatic increase in thymus weight, which was less pronounced in Rassf10 double knockout animals (Fig. 3b; Supplementary Fig. S5). The mean thymus weight (all weighed thymus) increased by 87% heterozygous Rassf10 knockout animals (344.8 mg) compared with wildtyp (184.6 mg). In addition, we observed that the median spleen weight increased in Rassf10+/− by 95% vs. Rassf10+/+ (Fig. 3d). We also found a more than threefold increased occurrence of kidney cysts in Rassf10 knockout (p53−/−) animals 25% Rassf10−/−, 11% Rassf10+/− vs. 8% wt (Table 2 and Fig. 3c, e). Kidney cysts were found in the medulla and cortex, which led to a compression and the structural loss of adjacent tissue, as well as size increase of the kidney when multiple cysts were present (Fig. 3e). Kidney cysts are similar to cancer with uncontrolled growth, the loss of tissue structure/function and loss of apoptotic control [36] and can be regarded a neoplasia [37].
a Kaplan–Meier survival curves for Rassf10 knockout (KO) mice reveal reduced survival vs. wt- Rassf10 animals in p53−/− background (not significant, log rank test). Number of animals are 24 Rassf10+/+, 77 Rassf10+/−/Rassf10−/−. b Heterozygous Rassf10 mice suffered from an increased thymus size/thymoma in comparison to wildtype Rassf10 mice in p53−/− background. Number of animals are 18 Rassf10+/+, 26 Rassf10+/−, and 33 Rassf10−/−. c The Rassf10 KO induces enlarged thymus (thymoma) and induces cystic kidneys. Tissues were isolated during necropsy, fixed with formaldehyde, paraffin embedded, 10 µm sections were prepared and H&E stained before microscopic analysis. d Median spleen weight in p53−/− background increases from Rassf10+/+ 100 mg (n = 5) to Rassf10+/− 195 mg (n = 10; Rassf10−/− 98.1 mg, n = 15). e Rassf10 knockout induces cystic kidneys in p53−/− background and found in kidney medulla and cortex (10-μm sections, 20×). Cysts (asterisk) and glomeruli (►) are indicated. Fifty micrometers standard is shown.
RASSF10 is epigenetically inactivated in human kidney cancer
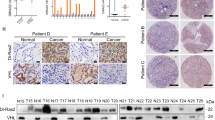
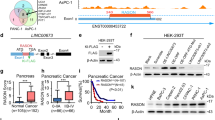
Tumor-suppressor inactivation in cancer can occur by the loss of function mutation or promoter methylation [38, 39]. TP53 is heavily mutated in various cancers especially in lung (>80%), head and neck (>70%), colorectal (>50%), breast (>30%), kidney chromophobe (30%) and below 5% in kidney clear cell carcinoma [40]. For RASSF10, there are no reported mutations and RASSF1 mutations are below 2% across primary cancers (Supplementary Table S1; TCGA; analyzed [41]). RASSF10 contains a CpG island in its promoter region (Supplementary Fig. S1) and is expressed in normal human kidney, lung, skin, brain, and colon (Fig. 4a). Given the high RASSF10 expression in the kidney and the affected kidneys upon Rassf10 knockout, we analyzed that the RASSF10 promoter methylation in human kidney cancer. We found that RASSF10 is epigenetically inactivated by promoter hypermethylation in 60% of kidney cancer cell lines (9/15, Fig. 4b). We could significantly reestablish the expression of RASSF10 by DNMT inhibition treatment with 5-Aza-2′deoxycytidine in MZ1257 and MZ1973 (Fig. 4c). Primary tumors of renal clear cell and renal papillary cell carcinoma confirmed that RASSF10 methylation correlated with decreased expression in comparison with normal kidney tissue (Fig. 4d, e). Using epigenetic editing by CRISPR-dCas9 system we were able to regulate RASSF10 expression (Fig. 4f, g). The targeted recruitment of epigenetic modulators/writers by the nuclease-deactivated-Cas9 to RASSF10 resulted in its expression modulation. Recruitment of histone acetyltransferase p300 activated RASSF10 and histone methyltransferase EZH2 reduced RASSF10 level in HEK293 cells (Fig. 4g). This result confirms the epigenetic regulation of RASSF10 in human kidney cells.
a RASSF10 expression across human normal tissues (log2, data GTEX—n = 2921—RPKM—ensgtexv4). b Kidney cancer cell lines were analyzed by combined bisulfite restriction analysis (+TaqI digested; −mock digest) for RASSF10 promoter hypermethylation and 9 out of 15 (60%) were methylated (m). HeLa was used as positive control. HEK293 cell line is partially methylated (pm). c Pharmacological inhibition of DNA-methyl-transferases by 5-Aza-2′deoxycytidine (Aza) significantly reestablished RASSF10 expression in kidney cancer cell lines MZ1257 and MZ1973 (t-test). RASSF10 was normalized to ß-ACTIN. d In kidney cancer patient samples RASSF10 methylation (cg05817758 in beta value) blocks RASSF10 expression (log2; norm. rsem + 1) shown for renal clear cell and e renal papillary carcinoma. Analyzed using [79]. f RASSF10 inhibition by CRISPR-Cas9 genomic targeting of RASSF10. HEK293 (RASSF10-TetOn) cells were transfected with CRISPR-Cas9 guide RNAs in px549 vector targeting RASSF10 (three guide combinations) for 48 h and RASSF10 was induced by doxycycline for 24 h. g RASSF10 is epigenetically regulated by EZH2 and p300. HEK293 (RASSF10-TetOn) cells were transfected with RASSF10 guide RNAs in px549 (deadCas) targeting RASSF10 and recruiting EZH2(deadCas) or p300(deadCas) for 72 h and RASSF10 was induced by doxycycline for 24 h. Protein lysates were separated by SDS-Page and western blotted with indicated antibodies.
RASSF10 inactivation correlates with clinical diagnosis and prognosis of human neoplasia of the kidney
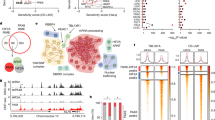
Next, we tested the ability of RASSF10 to serve as a prognostic and diagnostic biomarker in independent data sets for human neoplasia across various primary samples. RASSF10 expression is decreased in various types of kidney cancer (chromophobe, papillary, and clear cell, Fig. 5a), which was verified in a further data set and each was significant compared with normal kidney tissues (Fig. 5b). Reduced RASSF10 expression correlated with progressed tumor stage in renal chromophobe and renal clear cell carcinoma (Fig. 5c, d). In renal carcinoma (papillary and clear cells) overall survival correlated with high RASSF10 expression (Fig. 5e, f). We found that the low RASSF10 methylation levels are favorable for kidney cancer survival (renal papillary and clear cell carcinoma, Fig. 5g, h). Since we observed in the Rassf1A−/− and TP53−/− mice background increased neoplasia for the Rassf10 knockout (Table 2), we analyzed overall survival rate in 288 renal papillary carcinomas with the low RASSF10 and RASSF1 or TP53 expression in renal papillary cell carcinoma [42] (Supplementary Fig. S6). We observed that the low RASSF10 and low RASSF1 expression was significantly associated with impaired survival (p = 0.018), however, this was not significant for the low TP53 expression (Fig. S6).
a RASSF10 expression is decreased in various kidney cancer types (log2, data Kort, Anova one way) and RASSF10 downregulation was verified in b chromophobe, papillary and clear cell renal carcinoma (log2, data TCGA, Wilcoxon). c The loss of RASSF10 expression correlated with kidney tumor stage in chromophobe cell carcinoma (log2, data TCGA, Anova one way) and d in clear cell carcinoma (data TCGA, Anova one way). e In renal papillary cell carcinoma and f renal clear cell carcinoma low RASSF10 levels correlated with reduced 5-year overall survival (data Pan-Cancer RNAseq). High RASSF10 methylation levels correlated with reduced survival in g renal papillary and h clear cell carcinoma (data TCGA). i RASSF10 levels are reduced with increasing cyst size in polycystic kidney disease (ADPKD, small cysts: <1 ml with n = 5 and each pool of 4; medium cysts: 10–25 ml with n = 5; large cysts > 50 ml with n = 3), (log2, data Pei, Anova one way).
In polycystic kidney disease, there was a correlation between reduced RASSF10 expression and an increasing cyst size (Fig. 5i), consistent with Rassf10 loss driven renal cysts in mice and the longstanding idea of polycystic kidney disease as a neoplasia in disguise [37]. In summary, we observed that the levels of RASSF10 expression/methylation in combination with RASSF1A expression are suitable for prognosis and diagnosis of various kidney cancer types in humans.
Upregulation of KRAS signaling and MYC are associated with the loss of RASSF10 and impaired survival
To gain insight in the molecular processes that are deregulated upon Rassf10 inactivation we utilized mouse embryonic fibroblasts (MEFs) isolated from Rassf10 knockout and wildtype mice. Subsequently, we performed RNA microarray analysis to analyze the altered gene expression in knockout compared with wildtype MEFs. Gene ontology analysis of deregulated RNA levels was performed and this analysis shows a significant association with the molecular function of signaling receptor, molecular transducer and transmembrane signaling receptor activity (Table 3). Gene sets enrichment analysis showed a significant upregulation of IL6-JAK-STAT signaling and KRAS signaling in Rassf10 deficient MEFs compared with wildtype (Fig. 6a, b, respectively). Other hallmarks that were significantly associated the Rassf1a knockout belong to allograft rejection, inflammatory response, complement, and interferon gamma response gene sets. These results suggest that upon Rassf10 depletion several oncogenic pathways including RAS signaling are activated.
RNA isolated from Rassf10−/− and Rassf10 wildtype mouse embryonic fibroblasts were analyzed on MoGene-2_0-stRNA microarrays. Upregulated hallmarks were identified by gene set enrichment analysis. Enrichment plot for a IL6-JAK-STAT3 and b KRAS signaling are depicted. c MYC and d VEGFA are significantly negatively correlated with RASSF10 expression in papillary cell carcinoma (TCGA data set). Mean expression of RASSF10 and e MYC or f VEGFA in renal papillary cell carcinoma was analyzed in the pan-cancer RNA-seq panel with Kaplan–Meier plotter [42] and survival probability was plotted for low RASSF10 and high MYC/VEGF (black) expressing and high RASSF10 and low MYC/VEGF (red) expressing samples. g In clear cell carcinoma MYC is significantly negatively associated with RASSF10 expression and h expression of CDH1 is positively correlated with RASSF10. Correlation analysis of expression data using R2 Genomics Analysis and Visualization Platform [71] from human primary renal carcinomas (log2 data, TCGA data set).
MYC is an oncogenic downstream target of KRAS and IL6-JAK-STAT signaling [43, 44] and therefore we correlated the expression of MYC and RASSF10 in primary renal papillary cell carcinoma and clear cell carcinoma (Fig. 6c, g). In papillary cell carcinoma, we found a significant inverse correlation of RASSF10 and MYC or VEGF expression (Fig. 6c, p < 0.006 and Fig. 6d, p < 0.001, respectively). VEGF is an bona fide target of IL6-JAK-STAT pathway involved in angiogenesis [45]. Furthermore, we correlated overall survival of renal papillary cell carcinoma patients with RASSF10 and MYC or VEGF expression by Kaplan–Meier analysis (Fig. 6e, f). For both analysis a significant impaired survival was observed for low RASSF10 and high MYC or VEGF expression compared with the high RASSF10 and low MYC or VEGF (p = 1.6 × 10−4 and p = 2.1 × 10−7; Fig. 6e, f, respectively). In renal clear cell carcinoma, we also detected an inverse correlation between RASSF10 and MYC expression indicating that the loss of RASSF10 is associated with MYC induction (Fig. 6g, p < 0.004). Recently, it has been reported that RASSF10 regulates epithelial cadherin (CDH1) expression, that is a tumor suppressive TGFβ-target and inhibits epithelial-to-mesenchymal transition (EMT) [46]. Interestingly, we found that RASSF10 expression is positively associated with increased CDH1 levels in renal clear cell carcinoma (Fig. 6h; p = 4.6 × 10−9). Our data suggest that MYC and VEGF are negatively regulated by RASSF10 and that combination of high RASSF10 and low MYC or VEGF expression is associated with a favorable prognosis for renal cancer patients.
Discussion
We report for the first time that Rassf10 promotes tumor formation in vivo in a double knockout background with the p53 or Rassf1A tumor suppressors and this resulted in reduced survival and increased disease burden (Figs. 2 and 3). Rassf10 knockout promotes increased numbers of thymoma (p53−/− [47]), cystic kidneys (p53−/− [48, 49]), lymphoma (Rassf1A−/− [34], p53−/− [32, 50]), and splenomegaly (Rassf1A−/−, p53−/− [51]). Rassf10 heterozygous animals already suffer from an overall increased disease incidence (Table 2), suggesting Rassf10 is a haploinsufficient tumor suppressor, and the loss of one allele is sufficient for its loss of function and contribution to carcinogenesis. The effects could be directly attributable to the reduction in gene dosage or may act in concert with other oncogenic or haploinsufficient events (like PTEN, p53 or p27) [52, 53]. The mouse genetic background has a strong impact on tumor susceptibility and is mouse strain dependent [54, 55]. We have chosen the well characterized and established in house strain C57BL/6 for which Rassf1A and p53 knockouts existed and confirmed expression of Rassf10 in different tissues, including kidney (Fig. 1). Given the relative tumor suppressive influence of the C57BL/6 background [55,56,57], one could anticipate to observe further tumor types upon Rassf10 loss, like we report here for human patients, in other mouse strains or in combination with other tumor suppressors or even in an oncogenic background (e.g., KRAS), which might be addressed by others in the future. As a tumor suppressor in vivo, we wanted to understand the RASSF10 mechanism of regulation and contribution to carcinogenesis. We reported that knockdown of RASSF10 increased mitosis and increased cell proliferation and invasion, as well as RASSF10 reexpression halted proliferation [20, 26, 46]. In addition, RASSF10 was found at centrosomes/microtubules during mitosis [19]. Interestingly, we found that the loss of Rassf10 in MEFs is associated with upregulation of KRAS and IL6/JAK/STAT3 signaling (Fig. 6). The alteration of these oncogenic pathways is hallmarks in human carcinogenesis and leads to activation of signaling receptor and molecular transducer pathways, which was detected in the Rassf10 knockout MEFs (Table 3). RASSF10 contains a N-terminal Ras-association domain, but it is direct interaction which Ras has been not reported. A RASSF member, which contains a C-terminal RA domain and is bound to Ras, is the novel Ras effector NORE1 (RASSF5) [58]. It is interesting to note that mutations of Ras oncogenes do not play a major role in the initiation and progression of renal carcinomas [59]. This suggests that the activation of KRAS signaling may be accomplished by the inactivation of RASSF10.
Across different renal cancer entities, we found a significant epigenetic inactivation of RASSF10 (Figs. 4 and 5). In addition, we can exclude mutation events in the inactivation of RASSF10 in cancer (Table S1). DNA hypermethylation of gene promoters in cancer is a well-established mechanism of tumor-suppressor inactivation [60]. We could clearly show that the loss of RASSF10 expression in kidney cancer correlated with its increasing promoter methylation (Fig. 4). The successful pharmacological inhibition of DNA methylation restored RASSF10 expression, which we reported earlier in other cancer entities [20, 21, 26, 27]. We suggest that for the inactivation of the haploinsufficient tumor-suppressor RASSF10 the methylation of one allele is sufficient and found that low methylation levels of RASSF10 already decreased its expression dramatically. This is consistent with the observation that we neither found RASSF10 mutations in cancer nor LOH for the RASSF10 locus. Our clinical patient data revealed that the loss of RASSF10 is a common and general event in kidney carcinogenesis (Fig. 5). We could also show that in independent data sets RASSF10 loss correlated not only with reduced patient survival rates, but also with tumor stage/grade and was found in different kidney entities (clear cell carcinoma, papillary cell carcinoma, and chromophobe cell cancer). In addition, we observed that RASSF10 downregulation significantly correlates with upregulation of MYC or VEGF levels (Fig. 6). MYC and VEGF are bona fide oncogenic downstream targets of KRAS and IL6/JAK/STAT3 signaling pathways [43,44,45]. Low RASSF10 and high MYC or high VEGF was significantly associated with poor prognosis of renal papillary cell cancer patients and the combination of two markers had a higher impact on impaired probability of survival compared with the low RASSF10 alone (Figs. 5e and 6). Moreover, we found that RASSF10 expression is positively correlated with E-cadherin (CDH1) expression (Fig. 6). CDH1 is a tumor suppressive regulator of cell adhesion in epithelial cells and prevents EMT that is an important step in tumor invasion [61]. It has been reported that RASSF10 regulates CDH1 expression through the apoptosis stimulating protein of p53 (ASPP2) and inhibits TGFβ induced invasion of lung cancer cells [46].
Here we achieved the discovery and validation of the RASSF10 biomarker across different kidney tumor types, which in the next steps of biomarker development will have to be followed by assay development and analytical validation, clinical utility validation, and ultimately clinical implementation [2]. There are several FDA-approved cancer biomarkers currently used in clinical practice for e.g. liver, prostate, ovarian, breast, pancreatic, lung, and thyroid cancer [62, 63] and a blood-based colorectal DNA methylation marker screening test [64]. An epigenetic assay for RASSF1A methylation in prostate cancer, however, still biopsy based, is available [65, 66]. We could show that epigenetic editing of RASSF10 is possible, and one could think of targeted therapies in the future using virally applied CRISPR-Cas9 Epigenetic Editors to patients for reactivation of e.g. hypermethylated RASSF10 specifically in cancer (Fig. 4). RASSF10 is inactivated in various cancer types by DNA hypermethylation of its promoter. We are presenting compelling evidence that the haploinsufficient tumor-suppressor RASSF10 can be used as a prognostic and diagnostic cancer biomarker in combination with other tumor related genes (e.g., MYC) in kidney diseases.
Materials and methods
Rassf10 mouse
The bacterial artificial chromosome (BAC) backbone pBACe3.6 containing the single exon of Rassf10 (Source BioScience) was equipped with homologous regions 5′ and 3′ to Rassf10 genomic sequence for homologous recombination. Rassf10 cassette was then inserted into pKO24 vector and altered to contain LoxP sites flanking the coding region of Rassf10, as well as a neomycin cassette 3′ of lower LoxP site for positive selection. Vector contained diphtheria-toxin A cassette for negative selection. Completed Rassf10 knockout vector was used for homologous recombination in mouse embryonal stem cells of C57BL/6 mouse strain. Floxed Rassf10 containing animals (verified by southern blotting) were interbred with Cre recombinase expressing animals and the deletion of Rassf10 was verified by southern blotting, genomic PCR and on RNA expression levels.
Rassf10 mouse experimental setup
Rassf10 heterozygous parental animals were interbred to determine altered mendelian ratio of offspring (n = 348). Rassf10 knockout and wildtype mice were analyzed for spontaneous tumor formation by aging (Rassf10+/+ n = 27, Rassf10−/− n = 44). When reaching 80 weeks of age, mice were sacrificed and complete necropsy was performed. Rassf10 knockout animals were interbred with p53 or Rassf1A deficient animals and animals were obtained from Johnny Kim (Max-Planck Institute, Bad Nauheim, Germany, p53 knockout animals) and from Gerd Pfeifer [34] (City of Hope, CA, USA; Rassf1A knockout animals). Experimental setup in p53−/− was 20 weeks and in Rassf1A−/− was 80 weeks as published earlier and full necropsy was performed. Tissues were formaldehyde fixed, ethanol dehydrated, paraffin embedded, sectioned for analysis by H&E staining and microscopy. All experimental mouse work was approved by local authorities, animal care was in accordance with institutional guidelines and local authorities guidelines (Regierungspräsidium Darmstadt) and was performed at Max-Planck Institute, Bad Nauheim, Germany at Thomas Boettger and Thomas Braun laboratory.
Southern blotting
Genomic DNA for genotyping was isolated and digested with EcoRI (5′ probe detection) and BamHI (3′ probe detection). Digestion products were separated in 0.8% TAE gel and blotted onto nylon membrane, which was hybridized with 5′/3′ probes at 65 °C and finally analyzed by phosphor imager. For Rassf10 probe generation: genomic Rassf10 flanking regions were amplified by PCR, cloned into pJet1.2/blunt (Thermo Fisher Scientific), BglII digested and extracted from agarose gel. Probes were then radioactively labeled by a-P [10]-dCTP incorporation (Rediprime DNA Labeling System, Amersham) and purified by MicroSpin G-50 columns (GE Healthcare).
RNA in situ hybridization
At first mouse Rassf10 was cloned into pCMVTag1, linearized by digestion with ApaL1, Rassf10 was transcribed by T7 polymerase, and thereby Rassf10-RNA labeled with the RNA Labeling DIG Kit (Roche), and followed by Rassf10-RNA probe purification. C57BL/6 embryos were isolated, paraformaldehyde fixed, hybridized with Rassf10-RNA probe, and developed by BCIP/NBT (Sigma) [67, 68]. The stained embryos were then paraffin embedded and sectioned (10 µM) for analysis.
DNA methylation analysis
Promoter region of RASSF10 and Rassf10 was analyzed by CpG plot http://www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/ and both show CpG islands (Fig. S1). Primers for bisulfite treated DNA were designed to bind only fully converted DNA and amplify promoter region (Supplementary Table S2). DNA methylation of the RASSF10 promoter was analyzed by combined bisulfite restriction analysis (CoBRA) [21, 27]. The CoBRA PCR product for RASSF10 is 241 bp (TaqI sites at 49 + 140) or for nested 167 bp (TaqI site at 66 bp). DNA was isolated after proteinase K (Thermo Fisher Scientific) digest and extracted either with phenol/chloroform or by QIAamp DNA extraction kit (Qiagen), and concentrations were determined by UV-photospectrometery. For COBRA methylation analysis, 2 µg genomic DNA was bisulfite treated (12 µl 0.1 M hydroquinone, 208 µl 1.9 M sodium metabisulfite, and pH 5.5 with NaOH) and incubated over night at 50 °C. DNA was purified using MSB Spin PCRapace (STRATEC Molecular), eluted in 50 µl H2O and followed by 10 min incubation with 5 µl 3 M NaOH at 37 °C. DNA was then precipitated with 100% ethanol and 7.5 M ammonium acetate and resolved in 1 × TE buffer. The subsequent PCR product was digested with 5 U of TaqI (Thermo Fisher Scientific) 1 h at 65 °C and resolved on 2% TBE gel together with mock control and DNA ladder. In vitro methylation of genomic DNA was performed using CpG Methyltransferase M.SssI (NEB) according to manufacturer’s protocol.
RNA expression analysis
RNA was isolated from human cell culture or mouse primary tissues (homogenized using Bioruptor, Diagenode) using Isol-RNA lysis procedure (Trizol, Thermo Fisher Scientific). RNA was DNase (Thermo Fisher Scientific) treated and then reversely transcribed by MMLV (Promega). Quantitative RT–PCR was performed in triplicate with SYBR select (Thermo Fisher Scientific) using Rotor-Gene 3000 (Qiagen). For primers see Supplementary Table S2. RNA microarrays (MoGene-2_0-st, Thermo Fisher Scientific) were performed according to manufacturer’s protocol (P/N 703174 Rev. 2) with 200 ng of total RNA. Reagents/equipments were GeneChip® WT PLUS Reagent Kit, P/N: 902280; GeneChip® Hybridization, Wash, and Stain Kit P/N 900720, GeneChip Scanner, GeneChip Fluidics Station 450, GeneChip Hybridization Oven 640, Bioanalyzer 2100 (Agilent) and RNA600 NanoKit (Agilent). Microarray data are available in the ArrayExpress database (www.ebi.ac.uk) under accession number E-MTAB-8296.
Cell culture and treatment of cell lines
Cell lines were grown in appropriate medium (DMEM, RPMI) supplemented with 10% FCS, 1% penicillin/streptomycin under cell culture conditions (37 °C, 5% CO2). For 5-Aza-2′-deoxycytidine (Aza) treatment cells were split to 10% density and Aza was added with fresh medium on 4 consecutive days at working concentrations of 5, 10, and 20 µM before cell isolation. Cell lines were transfected for indicated time points using Polyethylenimmin (PEI, 4,9 mM, Sigma) for HEK293 cells. Doxycycline (Dox, Thermo Fisher Scientific) was dissolved in water and used for RASSF10 induction in HEK293 cells within the TetOn-TREx-system at working concentration of 2 µg/ml for 48 h [69].
Generation of stable RASSF10 inducible HEK293 cells
HEK293 stably expressing the Tet repressor under Blasticidine (5 µg/ml, Roth) were obtained from Thermo Fisher Scientific as a part of the TetOn-TREx-System, and were used as control cell line. These cells were transfected with the cloned RASSF10-pcDNA4ToMyc for stable insertion of Dox-inducible-RASSF10. Cells were selected for RASSF10 using Zeocin (500 µg/ml, Thermo Fisher Scientific).
Western blotting
Proteins were separated via SDS-PAGE and western blotted onto PVDF membrane (Immobilon) for antibody-based detection. Luminata Crescendo Western HRP substrate (Millipore) was used for detection at VersaDoc Imaging System (BioRad). The following antibodies were used: a-GAPDH (FL335, sc-25778 from Santa Cruz), a-RASSF10 (AP12444c-ev2020, Abgent), and HRP-coupled secondary antibodies anti-rabbit (sc2004, sc2357, Santa Cruz).
Genomic and epigenetic editing CRISPR-Cas9
To genomically delete RASSF10, we performed CRISPR-Cas9 targeted knockout. CRISPR-Cas9 vectors were obtained from Lienhard Schmitz (Giessen, Germany) and RASSF10 targeting oligos/guide RNAs were generated according to protocol [70]. Four RASSF10 knockout oligos were created to generate a frameshift and RASSF10 loss (three combinations) in px549 with wt-Cas9 (Supplementary Table S3). We transfected A549-cells with the CRISPR-Cas9 RASSF10 oligos and selected for positive clones by puromycin (1 µg/ml) for 3 days. Clones were expanded and the knockout was verified by PCR based amplification of the RASSF10 genomic region and RNA showing the deletion and was further verified by western blot. For the epigenetic editing of RASSF10, we created a deactivated Cas9 (px549dCas, by deletion) and cloned the RASSF10 targeting guide RNAs. EZH2 (Ezh2[SET]-dCas9 Addgene) and p300 (pcDNA-dCas9-p300 Core Addgene) were used as epigenetic modifiers upon cotransfection in HEK293 with guide RNAs(dCas).
Statistical analysis
Gene expression, promoter methylation correlation, and Kaplan–Meier calculations were performed using R2 Genomics Analysis and Visualization Platform [71], Wanderer [72], KM Plotter [42, 73,74,75], and MethSurv [76]. Gene ontology analysis of microarray data of differentially expressed genes (DEGs; TGFß DEG according to [77, 78]) was performed using Gene Ontology enRIchment anaLysis and visuaLizAtion tool Gorilla [79]. For further calculations, we used GraphPad (https://www.graphpad.com/quickcalcs/). Mendelian ratio alteration was determined by Chi square test. Knockout mice disease proneness was calculated by Fisher Exact Test. Student’s t test (unpaired; two tailed) was used for reexpression of RASSF10 under Aza treatment.
References
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86.
Goossens N, Nakagawa S, Sun X, Hoshida Y. Cancer biomarker discovery and validation. Transl Cancer Res. 2015;4:256–69.
Henry NL, Hayes DF. Cancer biomarkers. Mol Oncol. 2012;6:140–6.
Sauter ER. Reliable biomarkers to identify new and recurrent cancer. Eur J Breast Health. 2017;13:162–7.
Kornberg Z, Cooperberg MR, Spratt DE, Feng FY. Genomic biomarkers in prostate cancer. Transl Androl Urol. 2018;7:459–71.
Zamay TN, Zamay GS, Kolovskaya OS, Zukov RA, Petrova MM, Gargaun A, et al. Current and prospective protein biomarkers of lung cancer. Cancers. 2017;9:1–22.
Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–9.
Haag T, Herkt CE, Walesch SK, Richter AM, Dammann RH. The apoptosis associated tyrosine kinase gene is frequently hypermethylated in human cancer and is regulated by epigenetic mechanisms. Genes Cancer. 2014;5:365–74.
Haag T, Richter AM, Schneider MB, Jimenez AP, Dammann RH. The dual specificity phosphatase 2 gene is hypermethylated in human cancer and regulated by epigenetic mechanisms. BMC Cancer. 2016;16:49.
Kiehl S, Herkt SC, Richter AM, Fuhrmann L, El-Nikhely N, Seeger W, et al. ABCB4 is frequently epigenetically silenced in human cancers and inhibits tumor growth. Sci Rep. 2014;4:6899.
Richter AM, Kiehl S, Koger N, Breuer J, Stiewe T, Dammann RH. ZAR1 is a novel epigenetically inactivated tumour suppressor in lung cancer. Clin Epigenetics. 2017;9:60.
Schagdarsurengin U, Pfeifer GP, Dammann R. Frequent epigenetic inactivation of cystatin M in breast carcinoma. Oncogene. 2007;26:3089–94.
Steinmann K, Richter AM, Dammann RH. Epigenetic silencing of erythropoietin in human cancers. Genes Cancer. 2011;2:65–73.
Cheng YY, Jin HC, Chan MWY, Chu WK, Grusch M. Epigenetic biomarkers in cancer. Dis Markers. 2018;2018:4987103.
Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796:114–28.
Sherwood V, Recino A, Jeffries A, Ward A, Chalmers AD. The N-terminal RASSF family: a new group of Ras-association-domain-containing proteins, with emerging links to cancer formation. Biochem J. 2010;425:303–11.
Volodko N, Gordon M, Salla M, Ghazaleh HA, Baksh S. RASSF tumor suppressor gene family: biological functions and regulation. FEBS Lett. 2014;588:2671–84.
Hesson LB, Dunwell TL, Cooper WN, Catchpoole D, Brini AT, Chiaramonte R, et al. The novel RASSF6 and RASSF10 candidate tumour suppressor genes are frequently epigenetically inactivated in childhood leukaemias. Mol Cancer. 2009;8:42.
Hill VK, Underhill-Day N, Krex D, Robel K, Sangan CB, Summersgill HR, et al. Epigenetic inactivation of the RASSF10 candidate tumor suppressor gene is a frequent and an early event in gliomagenesis. Oncogene. 2010;30:978–89.
Richter AM, Walesch SK, Wurl P, Taubert H, Dammann RH. The tumor suppressor RASSF10 is upregulated upon contact inhibition and frequently epigenetically silenced in cancer. Oncogenesis. 2012;1:e18.
Schagdarsurengin U, Richter AM, Wohler C, Dammann RH. Frequent epigenetic inactivation of RASSF10 in thyroid cancer. Epigenetics. 2009;4:571–6.
Wang Y, Ma T, Bi J, Song B, Zhou Y, Zhang C, et al. RASSF10 is epigenetically inactivated and induces apoptosis in lung cancer cell lines. Biomed Pharmacother. 2014;68:321–6.
Wei Z, Chen X, Chen J, Wang W, Xu X, Cai Q. RASSF10 is epigenetically silenced and functions as a tumor suppressor in gastric cancer. Biochem Biophys Res Commun. 2013;432:632–7.
Richter AM, Zimmermann T, Haag T, Walesch SK, Dammann RH. Promoter methylation status of Ras-association domain family members in pheochromocytoma. Front Endocrinol. 2015;6:21.
Richter AM, Haag T, Walesch S, Herrmann-Trost P, Marsch WC, Kutzner H, et al. Aberrant promoter hypermethylation of RASSF family members in merkel cell carcinoma. Cancers. 2013;5:1566–76.
Richter AM, Walesch SK, Dammann RH. Aberrant promoter methylation of the tumour suppressor RASSF10 and its growth inhibitory function in breast cancer. Cancers. 2016;8:1–12.
Helmbold P, Richter AM, Walesch S, Skorokhod A, Marsch W, Enk A, et al. RASSF10 promoter hypermethylation is frequent in malignant melanoma of the skin but uncommon in nevus cell nevi. J Investig Dermatol. 2012;132:687–94.
Liu W, Wang J, Wang L, Qian C, Qian Y, Xuan H, et al. Ras-association domain family 10 acts as a novel tumor suppressor through modulating MMP2 in hepatocarcinoma. Oncogenesis. 2016;5:e237.
Fan C, Wang W, Jin J, Yu Z, Xin X. RASSF10 is epigenetically inactivated and suppresses cell proliferation and induces cell apoptosis by activating the p53 signalling pathway in papillary thyroid carcinoma cancer. Cell Physiol Biochem. 2017;41:1229–39.
Li X, Liang Q, Liu W, Zhang N, Xu L, Zhang X, et al. Ras association domain family member 10 suppresses gastric cancer growth by cooperating with GSTP1 to regulate JNK/c-Jun/AP-1 pathway. Oncogene. 2016;35:2453–64.
Loeb LA. Human cancers express a mutator phenotype: hypothesis, origin, and consequences. Cancer Res. 2016;76:2057–9.
Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr., Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–21.
Donehower LA. The p53-deficient mouse: a model for basic and applied cancer studies. Semin Cancer Biol. 1996;7:269–78.
Tommasi S, Dammann R, Zhang Z, Wang Y, Liu L, Tsark WM, et al. Tumor susceptibility of Rassf1a knockout mice. Cancer Res. 2005;65:92–98.
van der Weyden L, Happerfield L, Arends MJ, Adams DJ. Megaoesophagus in Rassf1a-null mice. Int J Exp Pathol. 2009;90:101–8.
Seeger-Nukpezah T, Geynisman DM, Nikonova AS, Benzing T, Golemis EA. The hallmarks of cancer: relevance to the pathogenesis of polycystic kidney disease. Nat Rev Nephrol. 2015;11:515–34.
Grantham JJ. Polycystic kidney disease: neoplasia in disguise. Am J Kidney Dis. 1990;15:110–6.
Chen YJ, Tang QB, Zou SQ. Inactivation of RASSF1A, the tumor suppressor gene at 3p21.3 in extrahepatic cholangiocarcinoma. World J Gastroenterol. 2005;11:1333–8.
Herman JG. Hypermethylation of tumor suppressor genes in cancer. Semin Cancer Biol. 1999;9:359–67.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–520.
Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227.
Morales JK, Falanga YT, Depcrynski A, Fernando J, Ryan JJ. Mast cell homeostasis and the JAK-STAT pathway. Genes Immun. 2010;11:599–608.
Vaseva AV, Blake DR, Gilbert TSK, Ng S, Hostetter G, Azam SH, et al. KRAS suppression-induced degradation of MYC is antagonized by a MEK5-ERK5 compensatory mechanism. Cancer Cell. 2018;34:807–822 e807.
Xu Q, Briggs J, Park S, Niu G, Kortylewski M, Zhang S, et al. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005;24:5552–60.
Richter AM, Kuster MM, Woods ML, Walesch SK, Gokyildirim MY, Krueger M, et al. RASSF10 is a TGFbeta-target that regulates ASPP2 and E-Cadherin expression and acts as tumor suppressor that is epigenetically downregulated in advanced cancer. Cancers. 2019;11:1–24.
Dudgeon C, Chan C, Kang W, Sun Y, Emerson R, Robins H, et al. The evolution of thymic lymphomas in p53 knockout mice. Genes Dev. 2014;28:2613–20.
Schonenberger D, Harlander S, Rajski M, Jacobs RA, Lundby AK, Adlesic M, et al. Formation of renal cysts and tumors in Vhl/Trp53-deficient mice requires HIF1alpha and HIF2alpha. Cancer Res. 2016;76:2025–36.
Van Bodegom D, Saifudeen Z, Dipp S, Puri S, Magenheimer BS, Calvet JP, et al. The polycystic kidney disease-1 gene is a target for p53-mediated transcriptional repression. J Biol Chem. 2006;281:31234–44.
Kuperwasser C, Hurlbut GD, Kittrell FS, Dickinson ES, Laucirica R, Medina D, et al. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am J Pathol. 2000;157:2151–9.
Gostissa M, Bianco JM, Malkin DJ, Kutok JL, Rodig SJ, Morse HC 3rd, et al. Conditional inactivation of p53 in mature B cells promotes generation of nongerminal center-derived B-cell lymphomas. Proc Natl Acad Sci USA. 2013;110:2934–9.
Inoue K, Fry EA. Haploinsufficient tumor suppressor genes. Adv Med Biol. 2017;118:83–122.
Santarosa M, Ashworth A. Haploinsufficiency for tumour suppressor genes: when you don’t need to go all the way. Biochim Biophys Acta. 2004;1654:105–22.
Doetschman T. Influence of genetic background on genetically engineered mouse phenotypes. Methods Mol Biol. 2009;530:423–33.
Rivera J, Tessarollo L. Genetic background and the dilemma of translating mouse studies to humans. Immunity. 2008;28:1–4.
Klopstock N, Katzenellenbogen M, Pappo O, Sklair-Levy M, Olam D, Mizrahi L, et al. HCV tumor promoting effect is dependent on host genetic background. PLoS ONE. 2009;4:e5025.
Puccini J, Dorstyn L, Kumar S. Genetic background and tumour susceptibility in mouse models. Cell Death Differ. 2013;20:964.
Vavvas D, Li X, Avruch J, Zhang XF. Identification of Nore1 as a potential Ras effector. J Biol Chem. 1998;273:5439–42.
Xie N, Zhou Y, Sun Q, Tang B. Novel epigenetic techniques provided by the CRISPR/Cas9 system. Stem Cells Int. 2018;2018:7834175.
Baylin S, Bestor TH. Altered methylation patterns in cancer cell genomes: cause or consequence? Cancer Cell. 2002;1:299–305.
Vogelmann R, Nguyen-Tat MD, Giehl K, Adler G, Wedlich D, Menke A. TGFbeta-induced downregulation of E-cadherin-based cell-cell adhesion depends on PI3-kinase and PTEN. J Cell Sci. 2005;118:4901–12.
Kirwan A, Utratna M, O’Dwyer ME, Joshi L, Kilcoyne M. Glycosylation-based serum biomarkers for cancer diagnostics and prognostics. Biomed Res Int. 2015;2015:490531.
Selleck MJ, Senthil M, Wall NR. Making meaningful clinical use of biomarkers. Biomark Insights. 2017;12:1177271917715236.
Issa IA, Noureddine M. Colorectal cancer screening: an updated review of the available options. World J Gastroenterol. 2017;23:5086–96.
Partin AW, Van Neste L, Klein EA, Marks LS, Gee JR, Troyer DA, et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J Urol. 2014;192:1081–7.
Stewart GD, Van Neste L, Delvenne P, Delree P, Delga A, McNeill SA, et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: results of the MATLOC study. J Urol. 2013;189:1110–6.
Sweetman D, Rathjen T, Jefferson M, Wheeler G, Smith TG, Wheeler GN, et al. FGF-4 signaling is involved in mir-206 expression in developing somites of chicken embryos. Dev Dyn. 2006;235:2185–91.
Wystub K, Besser J, Bachmann A, Boettger T, Braun T. miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLoS Genet. 2013;9:e1003793.
Jimenez AP, Traum A, Boettger T, Hackstein H, Richter AM, Dammann RH. The tumor suppressor RASSF1A induces the YAP1 target gene ANKRD1 that is epigenetically inactivated in human cancers and inhibits tumor growth. Oncotarget. 2017;8:88437–52.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.
Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–93.
Diez-Villanueva A, Mallona I, Peinado MA. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics Chromatin. 2015;8:22.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–31.
Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. 2013;8:e82241.
Szasz AM, Lanczky A, Nagy A, Forster S, Hark K, Green JE, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–33.
Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10:277–88.
Han SM, Ryu HM, Suh J, Lee KJ, Choi SY, Choi S, et al. Network-based integrated analysis of omics data reveal novel players of TGF-beta1-induced EMT in human peritoneal mesothelial cells. Sci Rep. 2019;9:1497.
Johansson J, Tabor V, Wikell A, Jalkanen S, Fuxe J. TGF-beta1-induced epithelial-mesenchymal transition promotes monocyte/macrophage properties in breast cancer cells. Front Oncol. 2015;5:3.
Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009;10:1–7.
Acknowledgements
We thank Sylvia Thomas at the MPI for performing the microarrays analysis, Richard Volckmann for his help with R2 data at AMC and Julia Weber and Helga Linder for cloning of the Rassf10 mouse knockout vector. Open access funding provided by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Richter, A.M., Woods, M.L., Küster, M.M. et al. RASSF10 is frequently epigenetically inactivated in kidney cancer and its knockout promotes neoplasia in cancer prone mice. Oncogene 39, 3114–3127 (2020). https://doi.org/10.1038/s41388-020-1195-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-1195-6
This article is cited by
-
Epigenetically silenced apoptosis-associated tyrosine kinase (AATK) facilitates a decreased expression of Cyclin D1 and WEE1, phosphorylates TP53 and reduces cell proliferation in a kinase-dependent manner
Cancer Gene Therapy (2022)
-
Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy
Molecular Cancer (2020)