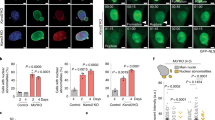
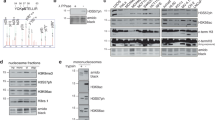
Abstract
Phosphofurin acidic cluster sorting protein-1 (PACS-1) is a multifunctional membrane traffic regulator that plays important roles in organ homeostasis and disease. In this study, we elucidate a novel nuclear function for PACS-1 in maintaining chromosomal integrity. PACS-1 progressively accumulates in the nucleus during cell cycle progression, where it interacts with class I histone deacetylases 2 and 3 (HDAC2 and HDAC3) to regulate chromatin dynamics by maintaining the acetylation status of histones. PACS-1 knockdown results in the proteasome-mediated degradation of HDAC2 and HDAC3, compromised chromatin maturation, as indicated by elevated levels of histones H3K9 and H4K16 acetylation, and, consequently, increased replication stress-induced DNA damage and genomic instability.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Gilbert N, Gilchrist S, Bickmore WA. Chromatin organization in the mammalian nucleus. Int Rev Cytol. 2005;242:283–336.
Peterson CL, Laniel M-A. Histones and histone modifications. Curr Biol. 2004;14:R546–51.
Leman AR, Noguchi E. The replication fork: understanding the eukaryotic replication machinery and the challenges to genome duplication. Genes. 2013;4:1–32.
Lande-Diner L, Zhang J, Cedar H. Shifts in replication timing actively affect histone acetylation during nucleosome reassembly. Mol Cell. 2009;34:767–74.
Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–41.
Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a018713.
Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–7.
Stengel KR, Hiebert SW. Class I HDACs affect DNA replication, repair, and chromatin structure: implications for cancer therapy. Antioxid Redox Signal. 2015;23:51–65.
Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun Z-W, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72.
Summers AR, Fischer MA, Stengel KR, Zhao Y, Kaiser JF, Wells CE, et al. HDAC3 is essential for DNA replication in hematopoietic progenitor cells. J Clin Invest. 2013;123:3112–23.
Wells CE, Bhaskara S, Stengel KR, Zhao Y, Sirbu B, Chagot B, et al. Inhibition of histone deacetylase 3 causes replication stress in cutaneous T cell lymphoma. PloS One. 2013;8:e68915.
Bhaskara S, Jacques V, Rusche JR, Olson EN, Cairns BR, Chandrasekharan MB. Histone deacetylases 1 and 2 maintain S-phase chromatin and DNA replication fork progression. Epigenetics Chromatin. 2013;6:27.
Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–32.
De Souza C, Chatterji BP. HDAC inhibitors as novel anti-cancer therapeutics. Recent Pat Anticancer Drug Disco. 2015;10:145–62.
Thomas G, Aslan JE, Thomas L, Shinde P, Shinde U, Simmen T. Caught in the act: protein adaptation and the expanding roles of the PACS proteins in tissue homeostasis and disease. J Cell Sci. 2017;130:1865–76.
Wan L, Molloy SS, Thomas L, Liu G, Xiang Y, Rybak SL, et al. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94:205–16.
Blagoveshchenskaya AD, Thomas L, Feliciangeli SF, Hung CH, Thomas G. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell. 2002;111:853–66.
Crump CM, Hung C-H, Thomas L, Wan L, Thomas G. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J Virol. 2003;77:11105–13.
Hinners I, Wendler F, Fei H, Thomas L, Thomas G, Tooze SA. AP-1 recruitment to VAMP4 is modulated by phosphorylation-dependent binding of PACS-1. EMBO Rep. 2003;4:1182–9.
Jenkins PM, Zhang L, Thomas G, Martens JR. PACS-1 mediates phosphorylation-dependent ciliary trafficking of the cyclic-nucleotide-gated channel in olfactory sensory neurons. J Neurosci J Soc Neurosci. 2009;29:10541–51.
Piguet V, Wan L, Borel C, Mangasarian A, Demaurex N, Thomas G, et al. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat Cell Biol. 2000;2:163–7.
Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM, et al. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem. 2007;282:32956–64.
Scott GK, Gu F, Crump CM, Thomas L, Wan L, Xiang Y, et al. The phosphorylation state of an autoregulatory domain controls PACS-1-directed protein traffic. EMBO J. 2003;22:6234–44.
Mendonca MS, Farrington DL, Mayhugh BM, Qin Y, Temples T, Comerford K, et al. Homozygous deletions within the 11q13 cervical cancer tumor-suppressor locus in radiation-induced, neoplastically transformed human hybrid cells. Genes Chromosomes Cancer. 2004;39:277–87.
Srivatsan ES, Bengtsson U, Manickam P, Benyamini P, Chandrasekharappa SC, Sun C, et al. Interstitial deletion of 11q13 sequences in HeLa cells. Genes Chromosomes Cancer. 2000;29:157–65.
Srivatsan ES, Chakrabarti R, Zainabadi K, Pack SD, Benyamini P, Mendonca MS, et al. Localization of deletion to a 300 Kb interval of chromosome 11q13 in cervical cancer. Oncogene. 2002;21:5631–42.
Zainabadi K, Jain AV, Donovan FX, Elashoff D, Rao NP, Murty VV, et al. One in four individuals of African-American ancestry harbors a 5.5kb deletion at chromosome 11q13.1. Genomics. 2014;103:276–87.
Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802.
Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–31.
Scott GK, Fei H, Thomas L, Medigeshi GR, Thomas G. A PACS-1, GGA3 and CK2 complex regulates CI-MPR trafficking. EMBO J. 2006;25:4423–35.
Crump CM, Xiang Y, Thomas L, Gu F, Austin C, Tooze SA, et al. PACS-1 binding to adaptors is required for acidic cluster motif-mediated protein traffic. EMBO J. 2001;20:2191–201.
Stern D, Cho MT, Chikarmane R, Willaert R, Retterer K, Kendall F, et al. Association of the missense variant p.Arg203Trp in PACS1 as a cause of intellectual disability and seizures. Clin Genet. 2017;92:221–3.
Gaillard H, García-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15:276–89.
Khurana S, Oberdoerffer P. Replication stress: a lifetime of epigenetic change. Genes. 2015;6:858–77.
Hayakawa T, Nakayama J. Physiological roles of class I HDAC complex and histone demethylase. J Biomed Biotechnol. 2011. https://doi.org/10.1155/2011/129383.
Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–47.
Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–9.
Hagelkruys A, Lagger S, Krahmer J, Leopoldi A, Artaker M, Pusch O, et al. A single allele of Hdac2 but not Hdac1 is sufficient for normal mouse brain development in the absence of its paralog. Dev Camb Engl. 2014;141:604–16.
Johnson CA, White DA, Lavender JS, O’Neill LP, Turner BM. Human class I histone deacetylase complexes show enhanced catalytic activity in the presence of ATP and co-immunoprecipitate with the ATP-dependent chaperone protein Hsp70. J Biol Chem. 2002;277:9590–7.
Ma P, Schultz RM. Histone Deacetylase 2 (HDAC2) regulates chromosome segregation and kinetochore function via H4K16 deacetylation during oocyte maturation in mouse. PLoS Genet. 2013;9. https://doi.org/10.1371/journal.pgen.1003377.
Mullican SE, Gaddis CA, Alenghat T, Nair MG, Giacomin PR, Everett LJ, et al. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. 2011;25:2480–8.
Wagner FF, Zhang Y-L, Fass DM, Joseph N, Gale JP, Weïwer M, et al. Kinetically selective inhibitors of histone deacetylase 2 (HDAC2) as cognition enhancers. Chem Sci. 2015;6:804–15.
Zhang X, Wharton W, Yuan Z, Tsai S-C, Olashaw N, Seto E. Activation of the growth-differentiation factor 11 gene by the histone deacetylase (HDAC) inhibitor trichostatin A and repression by HDAC3. Mol Cell Biol. 2004;24:5106–18.
Conti C, Leo E, Eichler GS, Sordet O, Martin MM, Fan A, et al. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70:4470–80.
Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–60.
Atkins KM, Thomas LL, Barroso-González J, Thomas L, Auclair S, Yin J, et al. The multifunctional sorting protein PACS-2 regulates SIRT1-mediated deacetylation of p53 to modulate p21-dependent cell-cycle arrest. Cell Rep. 2014;8:1545–57.
Godman CA, Joshi R, Tierney BR, Greenspan E, Rasmussen TP, Wang H-W, et al. HDAC3 impacts multiple oncogenic pathways in colon cancer cells with effects on Wnt and vitamin D signaling. Cancer Biol Ther. 2008;7:1570–80.
Senese S, Zaragoza K, Minardi S, Muradore I, Ronzoni S, Passafaro A, et al. Role for histone deacetylase 1 in human tumor cell proliferation. Mol Cell Biol. 2007;27:4784–95.
Yoon H-G, Chan DW, Huang Z-Q, Li J, Fondell JD, Qin J, et al. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–46.
Tripathi K, Mani C, Barnett R, Nalluri S, Bachaboina L, Rocconi RP, et al. Gli1 protein regulates the S-phase checkpoint in tumor cells via bid protein, and its inhibition sensitizes to DNA topoisomerase 1 inhibitors. J Biol Chem. 2014;289:31513–25.
Clark DW, Tripathi K, Dorsman JC, Palle K. FANCJ protein is important for the stability of FANCD2/FANCI proteins and protects them from proteasome and caspase-3 dependent degradation. Oncotarget. 2015;6:28816–32.
Tripathi K, Hussein UK, Anupalli R, Barnett R, Bachaboina L, Scalici J, et al. Allyl isothiocyanate induces replication-associated DNA damage response in NSCLC cells and sensitizes to ionizing radiation. Oncotarget. 2015;6:5237–52.
Tripathi K, Mani C, Clark DW, Palle K. Rad18 is required for functional interactions between FANCD2, BRCA2, and Rad51 to repair DNA topoisomerase 1-poisons induced lesions and promote fork recovery. Oncotarget. 2016;7:12537–53.
Acknowledgements
We thank Steve McClellan, Mitchell Cancer Institute for flow cytometry. This work is supported by grants from Abraham Mitchell Cancer Research Scholar Endowment grant, and partly by NIH grants R01GM098956 and R01CA219187 to KP, the NIH grants DK114855 and DK112844 to GT, the NIH grant R01GM108648 and DOD BRCP Breakthrough Award BC151728 to AV.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mani, C., Tripathi, K., Luan, S. et al. The multifunctional protein PACS-1 is required for HDAC2- and HDAC3-dependent chromatin maturation and genomic stability. Oncogene 39, 2583–2596 (2020). https://doi.org/10.1038/s41388-020-1167-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-1167-x
This article is cited by
-
iPSC-derived models of PACS1 syndrome reveal transcriptional and functional deficits in neuron activity
Nature Communications (2024)
-
Racial differences in RAD51 expression are regulated by miRNA-214-5P and its inhibition synergizes with olaparib in triple-negative breast cancer
Breast Cancer Research (2023)
-
Neural deficits in a mouse model of PACS1 syndrome are corrected with PACS1- or HDAC6-targeting therapy
Nature Communications (2023)
-
Integrating imaging-based classification and transcriptomics for quality assessment of human oocytes according to their reproductive efficiency
Journal of Assisted Reproduction and Genetics (2023)
-
PAX3/7-FOXO1 fusion-negative alveolar rhabdomyosarcoma in Schuurs-Hoeijmakers syndrome
Journal of Human Genetics (2022)