Abstract
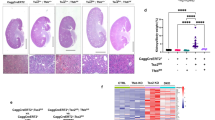
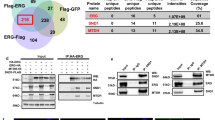
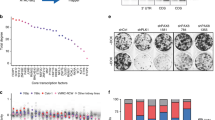
Tuberous sclerosis complex (TSC) is an autosomal dominant tumor suppressor syndrome, characterized by tumor development in multiple organs, including renal angiomyolipoma. Biallelic loss of TSC1 or TSC2 is a known genetic driver of angiomyolipoma development, however, whether an altered transcriptional repertoire contributes to TSC-associated tumorigenesis is unknown. RNA-seq analyses showed that MITF A isoform (MITF-A) was consistently highly expressed in angiomyolipoma, immunohistochemistry showed microphthalmia-associated transcription factor nuclear localization, and Chromatin immuno-Precipitation Sequencing analysis showed that the MITF-A transcriptional start site was highly enriched with H3K27ac marks. Using the angiomyolipoma cell line 621-101, MITF knockout (MITF.KO) and MITF-A overexpressing (MITF.OE) cell lines were generated. MITF.KO cells showed markedly reduced growth and invasion in vitro, and were unable to form xenografted tumors. In contrast, MITF.OE cells grew faster in vitro and as xenografted tumors compared to control cells. RNA-Seq analysis showed that both ID2 and Cysteine-rich angiogenic inducer 61 (CYR61) expression levels were increased in the MITF.OE cells and reduced in the MITF.KO cells, and luciferase assays showed this was due to transcriptional effects. Importantly, CYR61 overexpression rescued MITF.KO cell growth in vitro and tumor growth in vivo. These findings suggest that MITF-A is a transcriptional oncogenic driver of angiomyolipoma tumor development, acting through regulation of CYR61.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355:1345–56.
Henske EP, Jozwiak S, Kingswood JC, Sampson JR, Thiele EA. Tuberous sclerosis complex. Nat Rev Dis Primers. 2016;2:16035.
European Chromosome 16 Tuberous Sclerosis Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305–15.
van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Janssen B, Verhoef S, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805–8.
Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47:535–46.
Kim DH, Sarbassov dos D, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75.
Yecies JL, Manning BD. mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med. 2011;89:221–8.
Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513:440–3.
Giannikou K, Malinowska IA, Pugh TJ, Yan R, Tseng YY, Oh C, et al. Whole exome sequencing identifies TSC1/TSC2 biallelic loss as the primary and sufficient driver event for renal angiomyolipoma development. PLoS Genet. 2016;12:e1006242.
Martin KR, Zhou W, Bowman MJ, Shih J, Au KS, Dittenhafer-Reed KE, et al. The genomic landscape of tuberous sclerosis complex. Nat Commun. 2017;8:15816.
Carsillo T, Astrinidis A, Henske EP. Mutations in the tuberous sclerosis complex gene TSC2 are a cause of sporadic pulmonary lymphangioleiomyomatosis. Proc Natl Acad Sci USA. 2000;97:6085–90.
Badri KR, Gao L, Hyjek E, Schuger N, Schuger L, Qin W, et al. Exonic mutations of TSC2/TSC1 are common but not seen in all sporadic pulmonary lymphangioleiomyomatosis. Am J Respir Crit Care Med. 2013;187:663–5.
Bissler JJ, McCormack FX, Young LR, Elwing JM, Chuck G, Leonard JM, et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med. 2008;358:140–51.
McCormack FX, Inoue Y, Moss J, Singer LG, Strange C, Nakata K, et al. Efficacy and safety of sirolimus in lymphangioleiomyomatosis. N Engl J Med. 2011;364:1595–606.
Bonetti F, Chiodera PL, Pea M, Martignoni G, Bosi F, Zamboni G, et al. Transbronchial biopsy in lymphangiomyomatosis of the lung. HMB45 for diagnosis. Am J Surg Pathol. 1993;17:1092–102.
Zavala-Pompa A, Folpe AL, Jimenez RE, Lim SD, Cohen C, Eble JN, et al. Immunohistochemical study of microphthalmia transcription factor and tyrosinase in angiomyolipoma of the kidney, renal cell carcinoma, and renal and retroperitoneal sarcomas: comparative evaluation with traditional diagnostic markers. Am J Surg Pathol. 2001;25:65–70.
Hertzman Johansson C, Azimi A, Frostvik Stolt M, Shojaee S, Wiberg H, Grafstrom E, et al. Association of MITF and other melanosome-related proteins with chemoresistance in melanoma tumors and cell lines. Melanoma Res. 2013;23:360–5.
Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, et al. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–80.
Wellbrock C, Arozarena I. Microphthalmia-associated transcription factor in melanoma development and MAP-kinase pathway targeted therapy. Pigment Cell Melanoma Res. 2015;28:390–406.
Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;21:665–76.
Cheli Y, Ohanna M, Ballotti R, Bertolotto C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res. 2010;23:27–40.
Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–96.
GTEx Consortium. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–5.
Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19.
Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34.
Yu J, Astrinidis A, Howard S, Henske EP. Estradiol and tamoxifen stimulate LAM-associated angiomyolipoma cell growth and activate both genomic and nongenomic signaling pathways. Am J Physiol Lung Cell Mol Physiol. 2004;286:L694–700.
Fisher DE, Carr CS, Parent LA, Sharp PA. TFEB has DNA-binding and oligomerization properties of a unique helix-loop-helix/leucine-zipper family. Genes Dev. 1991;5:2342–52.
Harbst K, Staaf J, Lauss M, Karlsson A, Masback A, Johansson I, et al. Molecular profiling reveals low- and high-grade forms of primary melanoma. Clin Cancer Res. 2012;18:4026–36.
Hsiao JJ, Fisher DE. The roles of microphthalmia-associated transcription factor and pigmentation in melanoma. Arch Biochem Biophys. 2014;563:28–34.
Dhar A, Ray A. The CCN family proteins in carcinogenesis. Exp Oncol. 2010;32:2–9.
Lau LF. CCN1/CYR61: the very model of a modern matricellular protein. Cell Mol Life Sci. 2011;68:3149–63.
Mayer S, Erbes T, Timme-Bronsert S, Jaeger M, Rucker G, Kuf F, et al. Clinical relevance of Cyr61 expression in patients with hormone-dependent breast cancer. Oncol Lett. 2017;14:2334–40.
Tsai MS, Bogart DF, Castaneda JM, Li P, Lupu R. Cyr61 promotes breast tumorigenesis and cancer progression. Oncogene. 2002;21:8178–85.
Lin MT, Zuon CY, Chang CC, Chen ST, Chen CP, Lin BR, et al. Cyr61 induces gastric cancer cell motility/invasion via activation of the integrin/nuclear factor-kappaB/cyclooxygenase-2 signaling pathway. Clin Cancer Res. 2005;11:5809–20.
Gery S, Xie D, Yin D, Gabra H, Miller C, Wang H, et al. Ovarian carcinomas: CCN genes are aberrantly expressed and CCN1 promotes proliferation of these cells. Clin Cancer Res. 2005;11:7243–54.
Huang YT, Lan Q, Ponsonnet L, Blanquet M, Christofori G, Zaric J, et al. The matricellular protein CYR61 interferes with normal pancreatic islets architecture and promotes pancreatic neuroendocrine tumor progression. Oncotarget. 2016;7:1663–74.
Tong X, O’Kelly J, Xie D, Mori A, Lemp N, McKenna R, et al. Cyr61 suppresses the growth of non-small-cell lung cancer cells via the beta-catenin-c-myc-p53 pathway. Oncogene. 2004;23:4847–55.
Zhang H, Pasolli HA, Fuchs E. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc Natl Acad Sci USA. 2011;108:2270–5.
Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–33.
Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, et al. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–9.
Loercher AE, Tank EM, Delston RB, Harbour JW. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J Cell Biol. 2005;168:35–40.
Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22.
Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–39.
Li C, Lee PS, Sun Y, Gu X, Zhang E, Guo Y, et al. Estradiol and mTORC2 cooperate to enhance prostaglandin biosynthesis and tumorigenesis in TSC2-deficient LAM cells. J Exp Med. 2014;211:15–28.
Zarei M, Lal S, Parker SJ, Nevler A, Vaziri-Gohar A, Dukleska K, et al. Posttranscriptional upregulation of IDH1 by HuR establishes a powerful survival phenotype in pancreatic cancer cells. Cancer Res. 2017;77:4460–71.
Zarei M, Du H, Nassar AH, Yan RE, Giannikou K, Johnson SH, et al. Tumors with TSC mutations are sensitive to CDK7 inhibition through NRF2 and glutathione depletion. J Exp Med. 2019;216:2635–52.
Acknowledgements
This work was supported by the Engles Fund for TSC and LAM; the University of Pennsylvania Orphan Disease Center in partnership with the LAM Foundation; DOD-18-1-0592 (MZ), DOD-17-1-0205 (KG), and the NIH NCI 1P01CA120964 (DJK).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zarei, M., Giannikou, K., Du, H. et al. MITF is a driver oncogene and potential therapeutic target in kidney angiomyolipoma tumors through transcriptional regulation of CYR61. Oncogene 40, 112–126 (2021). https://doi.org/10.1038/s41388-020-01504-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-01504-8
This article is cited by
-
TFEB drives mTORC1 hyperactivation and kidney disease in Tuberous Sclerosis Complex
Nature Communications (2024)
-
A tissue-bioengineering strategy for modeling rare human kidney diseases in vivo
Nature Communications (2021)