Abstract
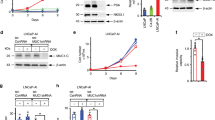
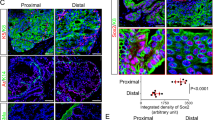
Neuroendocrine prostate cancer (NEPC) is an aggressive variant of prostate cancer that either develops de novo or arises from prostate adenocarcinoma as a result of treatment resistance. Although the prostate basal cells have been shown to directly generate tumor cells with neuroendocrine features when transduced with oncogenic signaling, the identity of the cell-of-origin for de novo NEPC remains unclear. We show that the TACSTD2high human prostate luminal epithelia cells highly express SOX2 and are relatively enriched in the transition zone prostate. Both TACSTD2high and TACSTD2low luminal cells transduced by constitutively activated AKT1 (caAKT1), and c-Myc can form organoids containing versatile clinically relevant tumor cell lineages with regard to the expression of AR and the neuroendocrine cell markers Synaptophysin and Chromogranin A. Tumor organoid cells derived from the TACSTD2high luminal cells are more predisposed to neuroendocrine differentiation along passaging and are relatively more castration-resistant. Knocking down TACSTD2 and SOX2 both attenuate neuroendocrine differentiation of tumor organoid cells. This study demonstrates de novo neuroendocrine differentiation of the human prostate luminal epithelial cells induced by caAKT1 and c-Myc and reveals an impact of cellular status on initiation of lineage plasticity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Conteduca V, Oromendia C, Eng KW, Bareja R, Sigouros M, Molina A, et al. Clinical features of neuroendocrine prostate cancer. Eur J Cancer. 2019;121:7–18.
Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78–83.
Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355:84–8.
Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305.
Lapuk AV, Wu C, Wyatt AW, McPherson A, McConeghy BJ, Brahmbhatt S, et al. From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer. J Pathol. 2012;227:286–97.
Dardenne E, Beltran H, Benelli M, Gayvert K, Berger A, Puca L, et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell. 2016;30:563–77.
Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–95.
Guo H, Ci X, Ahmed M, Hua JT, Soares F, Lin D, et al. ONECUT2 is a driver of neuroendocrine prostate cancer. Nat Commun. 2019;10:278.
Rotinen M, You S, Yang J, Coetzee SG, Reis-Sobreiro M, Huang WC, et al. ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis. Nat Med. 2018;24:1887–98.
Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, Kuruma H, et al. The master neural transcription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 2017;7:54–71.
Beltran H, Hruszkewycz A, Scher HI, Hildesheim J, Isaacs J, Yu EY, et al. The role of lineage plasticity in prostate cancer therapy resistance. Clin Cancer Res. 2019;25:6916–24.
Davies AH, Beltran H, Zoubeidi A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat Rev Urol. 2018;15:271–86.
Zou M, Toivanen R, Mitrofanova A, Floch N, Hayati S, Sun Y, et al. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 2017;7:736–49.
Lin D, Wyatt AW, Xue H, Wang Y, Dong X, Haegert A, et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 2014;74:1272–83.
Yao JL, Madeb R, Bourne P, Lei J, Yang X, Tickoo S, et al. Small cell carcinoma of the prostate: an immunohistochemical study. Am J Surg Pathol. 2006;30:705–12.
Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329:568–71.
Leong KG, Wang BE, Johnson L, Gao WQ. Generation of a prostate from a single adult stem cell. Nature. 2008;456:804–8.
Collins AT, Habib FK, Maitland NJ, Neal DE. Identification and isolation of human prostate epithelial stem cells based on alpha(2)beta(1)-integrin expression. J Cell Sci. 2001;114:3865–72.
Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci USA. 2007;104:181–6.
Lee JK, Phillips JW, Smith BA, Park JW, Stoyanova T, McCaffrey EF, et al. N-Myc drives neuroendocrine prostate Cancer initiated from human prostate epithelial cells. Cancer Cell. 2016;29:536–47.
Park JW, Lee JK, Sheu KM, Wang L, Balanis NG, Nguyen K, et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science. 2018;362:91–5.
Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–65.
Wang ZA, Mitrofanova A, Bergren SK, Abate-Shen C, Cardiff RD, Califano A, et al. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat Cell Biol. 2013;15:274–83.
Lu TL, Huang YF, You LR, Chao NC, Su FY, Chang JL, et al. Conditionally ablated Pten in prostate basal cells promotes basal-to-luminal differentiation and causes invasive prostate cancer in mice. Am J Pathol. 2013;182:975–91.
Liu J, Pascal LE, Isharwal S, Metzger D, Ramos Garcia R, Pilch J, et al. Regenerated luminal epithelial cells are derived from preexisting luminal epithelial cells in adult mouse prostate. Mol Endocrinol. 2011;25:1849–57.
Kwon OJ, Zhang L, Xin L. Stem cell antigen-1 identifies a distinct androgen-independent murine prostatic luminal cell lineage with bipotent potential. Stem Cells. 2016;34:191–202.
Karthaus WR, Hofree M, Choi D, Linton EL, Turkekul M, Bejnood A, et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science. 2020;368:497–505.
Kwon OJ, Choi JM, Zhang L, Jia D, Li Z, Zhang Y, et al. The Sca-1(+) and Sca-1(−) mouse prostatic luminal cell lineages are independently sustained. Stem Cells. 2020. https://doi.org/10.1002/stem.3253.
Joseph DB, Henry GH, Malewska A, Iqbal NS, Ruetten HM, Turco AE, et al. Urethral luminal epithelia are castration-insensitive cells of the proximal prostate. Prostate. 2020;80:872–84.
Crowell PD, Fox JJ, Hashimoto T, Diaz JA, Navarro HI, Henry GH, et al. Expansion of luminal progenitor cells in the aging mouse and human prostate. Cell Rep. 2019;28:1499–510.
Hsu EC, Rice MA, Bermudez A, Marques FJG, Aslan M, Liu S. et al. Trop2 is a driver of metastatic prostate cancer with neuroendocrine phenotype via PARP1. Proc Natl Acad Sci USA. Proc Natl Acad Sci USA. 2020;117:2032–42.
McNeal JE, Redwine EA, Freiha FS, Stamey TA. Zonal distribution of prostatic adenocarcinoma. Correlation with histologic pattern and direction of spread. Am J Surg Pathol. 1988;12:897–906.
Park JW, Lee JK, Phillips JW, Huang P, Cheng D, Huang J, et al. Prostate epithelial cell of origin determines cancer differentiation state in an organoid transformation assay. Proc Natl Acad Sci USA. 2016;113:4482–7.
Labrecque MP, Coleman IM, Brown LG, True LD, Kollath L, Lakely B, et al. Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J Clin Invest. 2019;130:4492–505.
Fine SW. Variants and unusual patterns of prostate cancer: clinicopathologic and differential diagnostic considerations. Adv Anat Pathol. 2012;19:204–16.
Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J. et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–75.
Xin L, Teitell MA, Lawson DA, Kwon A, Mellinghoff IK, Witte ON. Progression of prostate cancer by synergy of AKT with genotropic and nongenotropic actions of the androgen receptor. Proc Natl Acad Sci USA. 2006;103:7789–94.
Funding
This work was supported by R01CA190378, R01DK092202, and R01DK107436 (L.X.).
Author information
Authors and Affiliations
Contributions
Conception and design: LX and OK. Development of methodology: LX and OK. Acquisition of data: OK, LZ, DJ, ZZ, and ZL. Analysis and interpretation of data: OK, LZ, DJ, MH, and LX. Writing, review, and/or revision of the manuscript: LX and OK. Administrative, technical, or material support: LZ, DJ, LT, CM, and JKL, Study supervision: LX.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Kwon, OJ., Zhang, L., Jia, D. et al. De novo induction of lineage plasticity from human prostate luminal epithelial cells by activated AKT1 and c-Myc. Oncogene 39, 7142–7151 (2020). https://doi.org/10.1038/s41388-020-01487-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-01487-6
This article is cited by
-
Androgen-regulated stromal complement component 7 (C7) suppresses prostate cancer growth
Oncogene (2023)
-
Stromal FOXF2 suppresses prostate cancer progression and metastasis by enhancing antitumor immunity
Nature Communications (2022)
-
SOX2 mediates metabolic reprogramming of prostate cancer cells
Oncogene (2022)
-
Prostate luminal progenitor cells: from mouse to human, from health to disease
Nature Reviews Urology (2022)
-
Loss and revival of androgen receptor signaling in advanced prostate cancer
Oncogene (2021)