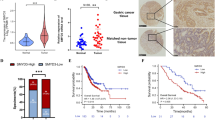
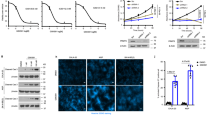
Abstract
Posttranslational modifications of histone and nonhistone proteins greatly influence numerous molecular events in multiple diseases. Jumonji domain-containing proteins are a family functioning as histone demethylase. Jumonji domain-containing protein 8 (JMJD8) is Jumonji C (JmjC) domain-only member of this family, and its physiological functions remain largely unknown. In this study, we investigated the mechanism by which aberrant JMJD8 stimulates phosphorylation of AKT and activate AKT/GSK3β/β-catenin signaling pathway thereby promotes tumor cell epithelial–mesenchymal transition (EMT). We demonstrated that knockdown of JMJD8 increased the interaction of SETDB1 and phosphoinositide-dependent kinase 1 (PDK1) with AKT1 and resulted in enhanced trimethylation of AKT1 at lysine 142 (K142), which is crucial for cell membrane recruitment, phosphorylation, and activation of AKT. Moreover, the mutation of histidine 200 of JMJD8 (JMJD8-H200Q) disrupted its binding with AKT1 and increased interaction of SETDB1 and PDK1 with AKT1. Furthermore, histone demethylase jumonji domain-containing protein 2B functioned as an adapter to recruit β-catenin to the methylated AKT1 upon JMJD8 depression, which facilitated the phosphorylation of β-catenin at Ser552 and its accumulation in cell nucleus where the activated β-catenin transcriptionally stimulated the expression of genes involved in EMT. In conclusion, our data unraveled a novel role of JMJD8 in regulating the migration and invasion of tumor via modulating AKT methylation and activation. In addition, this study showed that JMJD8 is a potential biomarker and drug design target for tumor EMT.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
26 March 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41388-021-01643-6
References
Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–70.
Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92.
Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27.
Kelly AD, Issa JJ. The promise of epigenetic therapy: reprogramming the cancer epigenome. Curr Opin Genet Dev. 2017;42:68–77.
Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–98.
Revathidevi S, Munirajan AK. Akt in cancer: mediator and more. Semin Cancer Biol. 2019;59:80–91.
De Santis MC, Gulluni F, Campa CC, Martini M, Hirsch E. Targeting PI3K signaling in cancer: challenges and advances. Biochim Biophys Acta Rev Cancer. 2019;1871:361–6.
Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta. 2011;1813:1938–45.
Hermida MA, Dinesh Kumar J, Leslie NR. GSK3 and its interactions with the PI3K/AKT/mTOR signalling network. Adv Biol Regul. 2017;65:5–15.
Zhang HH, Lipovsky AI, Dibble CC, Sahin M, Manning BD. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol Cell. 2006;24:185–97.
Song Y, Li ZX, Liu X, Wang R, Li LW, Zhang Q. The Wnt/beta-catenin and PI3K/Akt signaling pathways promote EMT in gastric cancer by epigenetic regulation via H3 lysine 27 acetylation. Tumour Biol. 2017;39:1010428317712617.
Jamieson C, Sharma M, Henderson BR. Wnt signaling from membrane to nucleus: beta-catenin caught in a loop. Int J Biochem Cell Biol. 2012;44:847–50.
Zhao L, Li W, Zang W, Liu Z, Xu X, Yu H, et al. JMJD2B promotes epithelial-mesenchymal transition by cooperating with beta-catenin and enhances gastric cancer metastasis. Clin Cancer Res. 2013;19:6419–29.
Yang F, Xu J, Li H, Tan M, Xiong X, Sun Y. FBXW2 suppresses migration and invasion of lung cancer cells via promoting beta-catenin ubiquitylation and degradation. Nat Commun. 2019;10:1382.
Yeo KS, Tan MC, Lim YY, Ea CK. JMJD8 is a novel endoplasmic reticulum protein with a JmjC domain. Sci Rep. 2017;7:15407.
Boeckel JN, Derlet A, Glaser SF, Luczak A, Lucas T, Heumuller AW, et al. JMJD8 regulates angiogenic sprouting and cellular metabolism by interacting with pyruvate kinase M2 in endothelial cells. Arterioscl Throm Vas. 2016;36(7):1425–33.
Yang Q, Jiang W, Hou P. Emerging role of PI3K/AKT in tumor-related epigenetic regulation. Semin Cancer Biol. 2019;59:112–24.
Guo J, Dai X, Laurent B, Zheng N, Gan W, Zhang J, et al. AKT methylation by SETDB1 promotes AKT kinase activity and oncogenic functions. Nat Cell Biol. 2019;21:226–37.
Wang G, Long J, Gao Y, Zhang W, Han F, Xu C, et al. SETDB1-mediated methylation of Akt promotes its K63-linked ubiquitination and activation leading to tumorigenesis. Nat Cell Biol. 2019;21:214–25.
Su Y, Wang J. JmjC domain-containing protein 8 (JMJD8) represses Ku70/Ku80 expression via attenuating AKT/NF-kappaB/COX-2 signaling. Biochim Biophys Acta Mol Cell Res. 2019;1866:118541.
Henderson V, Smith B, Burton LJ, Randle D, Morris M, Odero-Marah VA. Snail promotes cell migration through PI3K/AKT-dependent Rac1 activation as well as PI3K/AKT-independent pathways during prostate cancer progression. Cell Adh Migr. 2015;9:255–64.
Li Q, Hou L, Ding G, Li Y, Wang J, Qian B, et al. KDM6B induces epithelial-mesenchymal transition and enhances clear cell renal cell carcinoma metastasis through the activation of SLUG. Int J Clin Exp Pathol. 2015;8:6334–44.
Tang B, Qi G, Tang F, Yuan S, Wang Z, Liang X, et al. Aberrant JMJD3 expression upregulates slug to promote migration, invasion, and stem cell-like behaviors in hepatocellular carcinoma. Cancer Res. 2016;76:6520–32.
Tang B, Qi G, Tang F, Yuan S, Wang Z, Liang X, et al. JARID1B promotes metastasis and epithelial-mesenchymal transition via PTEN/AKT signaling in hepatocellular carcinoma cells. Oncotarget. 2015;6:12723–39.
Ghahhari NM, Babashah S. Interplay between microRNAs and WNT/beta-catenin signalling pathway regulates epithelial-mesenchymal transition in cancer. Eur J Cancer. 2015;51:1638–49.
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96.
Gao C, Xiao G, Hu J. Regulation of Wnt/beta-catenin signaling by posttranslational modifications. Cell Biosci. 2014;4:13.
Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–47.
Hino S, Tanji C, Nakayama KI, Kikuchi A. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol Cell Biol. 2005;25:9063–72.
Fang DX, Hawke D, Zheng YH, Xia Y, Meisenhelder J, Nika H, et al. Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. J Biol Chem. 2007;282:11221–9.
Kohn AD, Takeuchi F, Roth RA. Akt, a pleckstrin homology domain containing kinase, is activated primarily by phosphorylation. J Biol Chem. 1996;271:21920–6.
Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101.
Hamamoto R, Saloura V, Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer. 2015;15:110–24.
Guo J, Wei W. Fine-tuning AKT kinase activity through direct lysine methylation. Cell Cycle. 2019;18:917–22.
Mayer IA, Arteaga CL. The PI3K/AKT pathway as a target for cancer treatment. Annu Rev Med. 2016;67:11–28.
Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45.
Skrypek N, Goossens S, De Smedt E, Vandamme N, Berx G. Epithelial-to-mesenchymal transition: epigenetic reprogramming driving cellular plasticity. Trends Genet. 2017;33:943–59.
Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer. 2016;15:18.
Sun L, Fang J. Epigenetic regulation of epithelial-mesenchymal transition. Cell Mol Life Sci. 2016;73:4493–515.
Sechler M, Parrish JK, Birks DK, Jedlicka P. The histone demethylase KDM3A, and its downstream target MCAM, promote Ewing Sarcoma cell migration and metastasis. Oncogene. 2017;36:4150–60.
Yeo KS, Tan MC, Wong WY, Loh SW, Lam YL, Tan CL, et al. JMJD8 is a positive regulator of TNF-induced NF-kappaB signaling. Sci Rep. 2016;6:34125.
Alam H, Gu B, Lee MG. Histone methylation modifiers in cellular signaling pathways. Cell Mol Life Sci. 2015;72:4577–92.
Zhang J, Jing L, Li M, He L, Guo Z. Regulation of histone arginine methylation/demethylation by methylase and demethylase (review). Mol Med Rep. 2019;19:3963–71.
Oh S, Shin S, Janknecht R. The small members of the JMJD protein family: enzymatic jewels or jinxes? Biochim Biophys Acta Rev Cancer. 2019;1871:406–18.
Chu N, Salguero AL, Liu AZ, Chen Z, Dempsey DR, Ficarro SB, et al. Akt kinase activation mechanisms revealed using protein semisynthesis. Cell. 2018;174:897–907.e14.
Shin S, Janknecht R. Diversity within the JMJD2 histone demethylase family. Biochem Bioph Res Commun. 2007;353:973–7.
Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen ZZ, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–81.
Acknowledgements
This study was supported by the National Natural Science Foundation (11575232, 31471268), the National Key Research and Development Program of China (Stem Cell and Translational Research) 2016YFA0101202, the International Partnership Program of Chinese Academy of Sciences (116134KYSB20160084), and the Innovative Program of Development Foundation of Hefei Center for Physical Science and Technology (2016FXCX005).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Su, Y., Wang, X., Guo, Z. et al. Aberrant JmjC domain-containing protein 8 (JMJD8) expression promotes activation of AKT and tumor epithelial–mesenchymal transition. Oncogene 39, 6451–6467 (2020). https://doi.org/10.1038/s41388-020-01446-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-01446-1
This article is cited by
-
EGLN3 attenuates gastric cancer cell malignant characteristics by inhibiting JMJD8/NF-κB signalling activation independent of hydroxylase activity
British Journal of Cancer (2024)
-
Histone demethylases in the regulation of immunity and inflammation
Cell Death Discovery (2023)
-
JMJD family proteins in cancer and inflammation
Signal Transduction and Targeted Therapy (2022)