Abstract
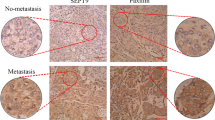
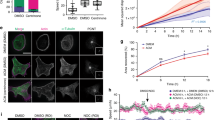
Cell motility is a tightly regulated phenomenon that supports the accurate formation of organ structure during development and homeostasis, including wound healing and inflammation. Meanwhile, cancer cells exhibit dysregulated motility, which causes spreading and invasion. The Dbl family RhoGEF ARHGEF7/β-PIX and its binding partner p21-activated kinase PAK1 are overexpressed in a variety of cancers and have been shown to be responsible for cancer cell migration. A key step in motility is the intracellular transport of ARHGEF7–PAK1 complex to the migrating front of cells, where lamellipodia protrusion and cytoskeletal remodeling efficiently occur. However, the molecular mechanisms of the intracellular transport of this complex are not fully understood. Here we revealed that SCL/TAL1-interrupting locus (STIL) is indispensable for the efficient migration of cancer cells. STIL forms a ternary complex with ARHGEF7 and PAK1 and accumulates with those proteins at the lamellipodia protrusion of motile cells. Knockdown of STIL impedes the accumulation of ARHGEF7–PAK1 complex within membrane ruffles and attenuates the phosphorylation of PAK1 substrates and cortical actin remodeling at the migrating front. Intriguingly, ARHGEF7 knockdown also diminishes STIL and PAK1 accumulation in membrane ruffles. Either STIL or ARHGEF7 knockdown impedes cell migration and Rac1 activity at the migrating front of cells. These results indicate that STIL is involved in the ARHGEF7-mediated positive-feedback activation of cytoskeletal remodeling through accumulating the ARHGEF7–PAK1 complex in lamellipodia. We conclude that its involvement is crucial for the polarized formation of Rac1-mediated leading edge, which supports the efficient migration of cancer cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Lawson CD, Ridley AJ. Rho GTPase signaling complexes in cell migration and invasion. J Cell Biol. 2018;217:447–57.
Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–90.
Koh CG, Manser E, Zhao ZS, Ng CP, Lim L. Beta1PIX, the PAK-interacting exchange factor, requires localization via a coiled-coil region to promote microvillus-like structures and membrane ruffles. J Cell Sci. 2001;114:4239–51.
Omelchenko T, Rabadan MA, Hernandez-Martinez R, Grego-Bessa J, Anderson KV, Hall A. beta-Pix directs collective migration of anterior visceral endoderm cells in the early mouse embryo. Genes Dev. 2014;28:2764–77.
Kumar R, Sanawar R, Li X, Li F. Structure, biochemistry, and biology of PAK kinases. Gene. 2017;605:20–31.
Rane CK, Minden A. P21 activated kinase signaling in cancer. Semin Cancer Biol. 2019;54:40–49.
Parrini MC, Lei M, Harrison SC, Mayer BJ. Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol Cell. 2002;9:73–83.
Vadlamudi RK, Li F, Adam L, Nguyen D, Ohta Y, Stossel TP, et al. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat Cell Biol. 2002;4:681–90.
Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, et al. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell 1998;1:183–92.
Byrne KM, Monsefi N, Dawson JC, Degasperi A, Bukowski-Wills JC, Volinsky N, et al. Bistability in the Rac1, PAK, and RhoA signaling network drives actin cytoskeleton dynamics and cell motility switches. Cell Syst. 2016;2:38–48.
Castro-Castro A, Ojeda V, Barreira M, Sauzeau V, Navarro-Lerida I, Muriel O, et al. Coronin 1A promotes a cytoskeletal-based feedback loop that facilitates Rac1 translocation and activation. EMBO J. 2011;30:3913–27.
Izraeli S, Lowe LA, Bertness VL, Good DJ, Dorward DW, Kirsch IR, et al. The SIL gene is required for mouse embryonic axial development and left-right specification. Nature 1999;399:691–4.
Patwardhan D, Mani S, Passemard S, Gressens P, El Ghouzzi V. STIL balancing primary microcephaly and cancer. Cell Death Dis. 2018;9:65.
Kasai K, Inaguma S, Yoneyama A, Yoshikawa K, Ikeda H. SCL/TAL1 interrupting locus derepresses GLI1 from the negative control of suppressor-of-fused in pancreatic cancer cell. Cancer Res 2008;68:7723–9.
Stevens NR, Dobbelaere J, Brunk K, Franz A, Raff JW. Drosophila Ana2 is a conserved centriole duplication factor. J Cell Biol. 2010;188:313–23.
Arquint C, Sonnen KF, Stierhof YD, Nigg EA. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J Cell Sci. 2012;125:1342–52.
Vulprecht J, David A, Tibelius A, Castiel A, Konotop G, Liu F, et al. STIL is required for centriole duplication in human cells. J Cell Sci. 2012;125:1353–62.
Nigg EA, Holland AJ. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat Rev Mol Cell Biol. 2018;19:297–312.
Erez A, Perelman M, Hewitt SM, Cojacaru G, Goldberg I, Shahar I, et al. Sil overexpression in lung cancer characterizes tumors with increased mitotic activity. Oncogene. 2004;23:5371–7.
Rabinowicz N, Mangala LS, Brown KR, Checa-Rodriguez C, Castiel A, Moskovich O, et al. Targeting the centriolar replication factor STIL synergizes with DNA damaging agents for treatment of ovarian cancer. Oncotarget. 2017;8:27380–92.
Wu X, Xiao Y, Yan W, Ji Z, Zheng G. The human oncogene SCL/TAL1 interrupting locus (STIL) promotes tumor growth through MAPK/ERK, PI3K/Akt and AMPK pathways in prostate cancer. Gene 2018;686:220–7.
Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell 2009;16:259–66.
Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS ONE. 2010;5:e13091.
Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 2006;9:121–32.
Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr Biol 2005;15:1276–85.
Schnoor M, Stradal TE, Rottner K. Cortactin: cell functions of a multifaceted actin-binding protein. Trends Cell Biol. 2018;28:79–98.
Yamao M, Naoki H, Kunida K, Aoki K, Matsuda M, Ishii S. Distinct predictive performance of Rac1 and Cdc42 in cell migration. Sci Rep. 2015;5:17527.
Bustelo XR, Ojeda V, Barreira M, Sauzeau V, Castro-Castro A. Rac-ing to the plasma membrane: the long and complex work commute of Rac1 during cell signaling. Small GTPases. 2012;3:60–6.
Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98:15149–54.
Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–10.
Michiels F, Habets GG, Stam JC, van der Kammen RA, Collard JG. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338–40.
Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–91.
Kurokawa K, Itoh RE, Yoshizaki H, Nakamura YO, Matsuda M. Coactivation of Rac1 and Cdc42 at lamellipodia and membrane ruffles induced by epidermal growth factor. Mol Biol Cell. 2004;15:1003–10.
Kunida K, Matsuda M, Aoki K. FRET imaging and statistical signal processing reveal positive and negative feedback loops regulating the morphology of randomly migrating HT-1080 cells. J Cell Sci. 2012;125:2381–92.
Schmidt M, Böhm D, von Törne C, Steiner E, Puhl A, Pilch H, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–13.
Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15:7642–51.
Aplan PD, Lombardi DP, Ginsberg AM, Cossman J, Bertness VL, Kirsch IR. Disruption of the human SCL locus by “illegitimate” V-(D)-J recombinase activity. Science. 1990;250:1426–9.
Nola S, Sebbagh M, Marchetto S, Osmani N, Nourry C, Audebert S, et al. Scrib regulates PAK activity during the cell migration process. Hum Mol Genet. 2008;17:3552–65.
Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, et al. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat Cell Biol. 2004;6:656–64.
Ohta M, Ashikawa T, Nozaki Y, Kozuka-Hata H, Goto H, Inagaki M, et al. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat Commun. 2014;5:5267.
Zhao ZS, Lim JP, Ng YW, Lim L, Manser E. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol Cell. 2005;20:237–49.
Li F, Adam L, Vadlamudi RK, Zhou H, Sen S, Chernoff J, et al. p21-activated kinase 1 interacts with and phosphorylates histone H3 in breast cancer cells. EMBO Rep. 2002;3:767–73.
Rosario CO, Kazazian K, Zih FS, Brashavitskaya O, Haffani Y, Xu RS, et al. A novel role for Plk4 in regulating cell spreading and motility. Oncogene. 2015;34:3441–51.
Kazazian K, Go C, Wu H, Brashavitskaya O, Xu R, Dennis JW, et al. Plk4 promotes cancer invasion and metastasis through Arp2/3 complex regulation of the actin cytoskeleton. Cancer Res 2017;77:434–47.
Acknowledgements
We thank Norika Yamada, Kazuko Tanimizu, and Minako Suzuki for their excellent technical assistance and Tatsuhito Miyake and Yuji Nakagomi for laser confocal microscope. We also thank Yukiko Kuru (Faculty of Foreign Languages, Aichi Medical University School of Medicine) for English editing. This study was supported in part by JSPS KAKENHI Grant Number 16K19091 (HI), 17K08706 (SI), 19K06671 (AI), 18K07031 (HM), 16H06280 “ABiS” (MM), 15H05949 “Resonance Bio” (MM), 24590456 and 19K07449 (KK); Pancreas Research Foundation of Japan (HI).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ito, H., Tsunoda, T., Riku, M. et al. Indispensable role of STIL in the regulation of cancer cell motility through the lamellipodial accumulation of ARHGEF7–PAK1 complex. Oncogene 39, 1931–1943 (2020). https://doi.org/10.1038/s41388-019-1115-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-1115-9
This article is cited by
-
ncRNAs-mediated overexpression of STIL predict unfavorable prognosis and correlated with the efficacy of immunotherapy of hepatocellular carcinoma
Cancer Cell International (2023)
-
Beta-Pix-dynamin 2 complex promotes colorectal cancer progression by facilitating membrane dynamics
Cellular Oncology (2021)