Abstract
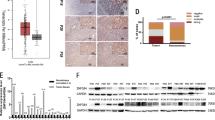
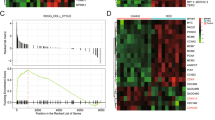
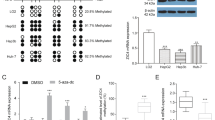
Discerning oncogenic drivers from passengers remain a major effort in understanding of the essence of the initiation and development of hepatocellular carcinoma (HCC), which is the most common primary liver malignancy and the third leading cause of cancer mortality worldwide. Here we report that ZNF774, a novel zinc-finger protein, inhibits the proliferation and invasion of HCC cells. Molecular characterization of this protein indicated that ZNF774 acts as a transcription repressor, and interrogation of ZNF774 interactome by affinity purification-coupled mass spectrometry revealed that ZNF774 is physically associated with the Mi-2/nucleosome remodeling and deacetylase (NuRD) complex in cells. Genome-wide identification of the transcriptional targets of the ZNF774/NuRD complex by ChIP-seq indicated that ZNF774 represses a cohort of genes including NOTCH2 that are critically involved in the growth and mobility of HCC. We demonstrated that the ZNF774/NuRD complex inhibits the proliferation and invasion of HCC cells in vitro and suppresses HCC growth and metastasis in vivo. Importantly, the expression of ZNF774 is significantly downregulated in HCC, and low ZNF774 expression strongly correlated with high NOTCH2 expression, advanced pathological stages, and poor overall survival of the patients. Together, these results uncover a key role for the ZNF774/NuRD-NOTCH2 axis in hepatocarcinogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
03 February 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41388-020-1180-0
References
El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. https://doi.org/10.1053/j.gastro.2007.2504.2061.
Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–14. https://doi.org/10.1016/S0140-6736(1318)30010-30012.
Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–3. https://doi.org/10.1038/nature09999.
Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64:2746–62. https://doi.org/10.1007/s00018-00007-07164-00011.
Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33. https://doi.org/10.1016/j.cell.2009.1003.1045.
McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M, et al. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development. 2001;128:491–502.
Serra H, Chivite I, Angulo-Urarte A, Soler A, Sutherland JD, Arruabarrena-Aristorena A, et al. PTEN mediates Notch-dependent stalk cell arrest in angiogenesis. Nat Commun. 2015;6:7935. https://doi.org/10.1038/ncomms8935.
Suzuki IK, Gacquer D, Van Heurck R, Kumar D, Wojno M, Bilheu A, et al. Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell. 2018;173:1370–.e1316. https://doi.org/10.1016/j.cell.2018.1303.1067.
Xiao W, Gao Z, Duan Y, Yuan W, Ke Y. Notch signaling plays a crucial role in cancer stem-like cells maintaining stemness and mediating chemotaxis in renal cell carcinoma. J Exp Clin Cancer Res. 2017;36:41. https://doi.org/10.1186/s13046-13017-10507-13043.
Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6.
Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–40. https://doi.org/10.1242/jcs.127308.
Dill MT, Tornillo L, Fritzius T, Terracciano L, Semela D, Bettler B, et al. Constitutive Notch2 signaling induces hepatic tumors in mice. Hepatology. 2013;57:1607–19. https://doi.org/10.1002/hep.26165.
Geisler F, Nagl F, Mazur PK, Lee M, Zimber-Strobl U, Strobl LJ, et al. Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology. 2008;48:607–16. https://doi.org/10.1002/hep.22381.
Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 2017;97:1235–94. https://doi.org/10.1152/physrev.00005.02017.
Wang J, Dong M, Xu Z, Song X, Zhang S, Qiao Y, et al. Notch2 controls hepatocyte-derived cholangiocarcinoma formation in mice. Oncogene. 2018;37:3229–42. https://doi.org/10.1038/s41388-41018-40188-41381.
Huntzicker EG, Hotzel K, Choy L, Che L, Ross J, Pau G, et al. Differential effects of targeting Notch receptors in a mouse model of liver cancer. Hepatology. 2015;61:942–52.
Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–11.
Totaro A, Castellan M, Di Biagio D, Piccolo S. Crosstalk between YAP/TAZ and Notch Signaling. Trends Cell Biol. 2018;28:560–73.
Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46:1267–73.
Huntzicker EG, Hotzel K, Choy L, Che L, Ross J, Pau G, et al. Differential effects of targeting Notch receptors in a mouse model of liver cancer. Hepatology. 2015;61:942–52. https://doi.org/10.1002/hep.27566.
Hayashi T, Gust KM, Wyatt AW, Goriki A, Jager W, Awrey S, et al. Not all NOTCH Is created equal: the oncogenic role of NOTCH2 in bladder cancer and its implications for targeted therapy. Clin Cancer Res. 2016;22:2981–92. https://doi.org/10.1158/1078-0432.CCR-2915-2360.
Hubmann R, Sieghart W, Schnabl S, Araghi M, Hilgarth M, Reiter M, et al. Gliotoxin targets nuclear NOTCH2 in human solid tumor derived cell lines in vitro and inhibits melanoma growth in xenograft mouse model. Front Pharm. 2017;8:319. https://doi.org/10.3389/fphar.2017.00319.
Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–7. https://doi.org/10.1038/nature08878.
Beel AJ, Sanders CR. Substrate specificity of gamma-secretase and other intramembrane proteases. Cell Mol Life Sci. 2008;65:1311–34. https://doi.org/10.1007/s00018-00008-07462-00012.
Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–83. https://doi.org/10.1038/embor.2008.1037.
Shih IeM, Wang TL. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67:1879–82. https://doi.org/10.1158/0008-5472.CAN-1806-3958.
van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–63. https://doi.org/10.1038/nature03659.
Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. https://doi.org/10.1186/gb-2003-1184-1110-1231.
Gebelein B, Fernandez-Zapico M, Imoto M, Urrutia R. KRAB-independent suppression of neoplastic cell growth by the novel zinc finger transcription factor KS1. J Clin Invest. 1998;102:1911–9. https://doi.org/10.1172/JCI1919.
Knight RD, Shimeld SM. Identification of conserved C2H2 zinc-finger gene families in the Bilateria. Genome Biol. 2001;2:RESEARCH0016.
Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18:128–34. https://doi.org/10.1038/nrc.2017.1118.
Lovisa S, LeBleu VS, Tampe B, Sugimoto H, Vadnagara K, Carstens JL, et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med. 2015;21:998–1009. https://doi.org/10.1038/nm.3902.
Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21–45. https://doi.org/10.1016/j.cell.2016.1006.1028.
Ong HT, Federspiel MJ, Guo CM, Ooi LL, Russell SJ, Peng KW, et al. Systemically delivered measles virus-infected mesenchymal stem cells can evade host immunity to inhibit liver cancer growth. J Hepatol. 2013;59:999–1006. https://doi.org/10.1016/j.jhep.2013.1007.1010.
Si W, Huang W, Zheng Y, Yang Y, Liu X, Shan L, et al. Dysfunction of the reciprocal feedback loop between GATA3- and ZEB2-nucleated repression programs contributes to breast cancer metastasis. Cancer Cell. 2015;27:822–36. https://doi.org/10.1016/j.ccell.2015.1004.1011.
Jung KH, Zhang J, Zhou C, Shen H, Gagea M, Rodriguez-Aguayo C, et al. Differentiation therapy for hepatocellular carcinoma: multifaceted effects of miR-148a on tumor growth and phenotype and liver fibrosis. Hepatology. 2016;63:864–79. https://doi.org/10.1002/hep.28367.
Ringelhan M, Pfister D, O’Connor T, Pikarsky E, Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19:222–32. https://doi.org/10.1038/s41590-41018-40044-z.
Lorsch ZS, Hamilton PJ, Ramakrishnan A, Parise EM, Salery M, Wright WJ, et al. Stress resilience is promoted by a Zfp189-driven transcriptional network in prefrontal cortex. Nat Neurosci. 2019;22:1413–23. https://doi.org/10.1038/s41593-41019-40462-41598.
Geisler F, Strazzabosco M. Emerging roles of Notch signaling in liver disease. Hepatology. 2015;61:382–92.
Cai Q, Zhang B, Sung H, Low SK, Kweon SS, Lu W, et al. Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. Nat Genet. 2014;46:886–90. https://doi.org/10.1038/ng.3041.
Capra JA, Erwin GD, McKinsey G, Rubenstein JL, Pollard KS. Many human accelerated regions are developmental enhancers. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130025. https://doi.org/10.1098/rstb.2013.0025.
Krushkal J, Ferrell R, Mockrin SC, Turner ST, Sing CF, Boerwinkle E. Genome-wide linkage analyses of systolic blood pressure using highly discordant siblings. Circulation. 1999;99:1407–10.
Goriki A, Seiler R, Wyatt AW, Contreras-Sanz A, Bhat A, Matsubara A, et al. Unravelling disparate roles of NOTCH in bladder cancer. Nat Rev Urol. 2018;15:345–57. https://doi.org/10.1038/s41585-41018-40005-41581.
Totaro A, Castellan M, Di Biagio D, Piccolo S. Crosstalk between YAP/TAZ and Notch signaling. Trends Cell Biol. 2018;28:560–73. https://doi.org/10.1016/j.tcb.2018.1003.1001.
Wu N, Nguyen Q, Wan Y, Zhou T, Venter J, Frampton GA, et al. The Hippo signaling functions through the Notch signaling to regulate intrahepatic bile duct development in mammals. Lab Invest. 2017;97:843–53. https://doi.org/10.1038/labinvest.2017.1029.
He L, Liu X, Yang J, Li W, Liu S, Liu X, et al. Imbalance of the reciprocally inhibitory loop between the ubiquitin-specific protease USP43 and EGFR/PI3K/AKT drives breast carcinogenesis. Cell Res. 2018;28:934–51. https://doi.org/10.1038/s41422-41018-40079-41426.
Li W, Zhang Z, Liu X, Cheng X, Zhang Y, Han X, et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J Clin Invest. 2017;127:3421–40. https://doi.org/10.1172/JCI94233.
Acknowledgements
This paper was supported by Capital Health Research and Special Development (No. 2016-1-1111), National Key Technologies R&D Program (No. 2015BAI13B09), Beijing Municipal Administration of Hospitals Incubator Program (No. PX2016001 and No. PX2019004) and “Miaopu” Innovation Foundation of the Chinese PLA General Hospital (No. 17KMM07).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Guan, C., He, L., Chang, Z. et al. ZNF774 is a potent suppressor of hepatocarcinogenesis through dampening the NOTCH2 signaling. Oncogene 39, 1665–1680 (2020). https://doi.org/10.1038/s41388-019-1075-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-1075-0
This article is cited by
-
The cancer-testis lncRNA LINC01977 promotes HCC progression by interacting with RBM39 to prevent Notch2 ubiquitination
Cell Death Discovery (2023)
-
Structures and biological functions of zinc finger proteins and their roles in hepatocellular carcinoma
Biomarker Research (2022)
-
Post-treatment cell-free DNA as a predictive biomarker in molecular-targeted therapy of hepatocellular carcinoma
Journal of Gastroenterology (2021)
-
Integrative analysis of mRNA and miRNA sequencing data for gliomas of various grades
Egyptian Journal of Medical Human Genetics (2020)