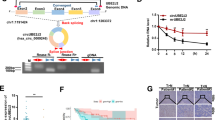
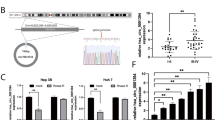
Abstract
Circular RNAs (circRNAs) are a novel class of RNAs, which involve in many physiological processes and participate in many diseases, especially in cancer. Previous reports showed circRNAs were globally downregulated in hepatocellular carcinoma (HCC) and a lot of circRNAs involved in the tumorigenesis and metastasis of HCC. To understand the underlying mechanism of circRNAs’ reduction, we explored the relationship between circRNA biogenesis and NUDT21, which was a RNA splice factor downregulated in HCC, and we found that NUDT21 elevated the formation of circRNA, and the UGUA sequences were critical for the cyclization of circRNA. Knockdown of NUDT21 disrupted the competitive endogenous RNA (ceRNA) pathway of circRNA-miRNA-mRNA, and overexpression of the downregulated circRNA could assist the NUDT21-mediated tumor suppression in HCC cells. In conclusion, the loss of NUDT21 prevented the cyclization of circRNA in HCC; without circRNA absorption, miRNAs were released to suppress the tumor-suppressor genes, leading to the uncontrolled cell proliferation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
El-Serag HB. Hepatocellular carcinoma. New Engl J Med. 2011;365:1118–27.
Zhang H, Sheng C, Yin Y, Wen S, Yang G, Cheng Z, et al. PABPC1 interacts with AGO2 and is responsible for the microRNA mediated gene silencing in high grade hepatocellular carcinoma. Cancer Lett. 2015;367:49–57.
Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38.
Ding J, Zhou W, Li X, Sun M, Ding J, Zhu Q. Tandem DNAzyme for double digestion: a new tool for circRNA suppression. Biol Chem. 2019;400:247–53.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–1.
Meng X, Chen Q, Zhang P, Chen M. CircPro: an integrated tool for the identification of circRNAs with protein-coding potential. Bioinformatics. 2017;33:3314–6.
Xu S, Zhou L, Ponnusamy M, Zhang L, Dong Y, Zhang Y, et al. A comprehensive review of circRNA: from purification and identification to disease marker potential. PeerJ. 2018;6:e5503.
Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–83.
Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057.
Song C, Li D, Liu H, Sun H, Liu Z, Zhang L, et al. The competing endogenous circular RNA ADAMTS14 suppressed hepatocellular carcinoma progression through regulating microRNA-572/regulator of calcineurin 1. J Cell Physiol. 2019;234:2460–70.
Zhang XW, Luo P, Jing W, Zhou H, Liang CZ, Tu JC. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. Oncotargets Ther. 2018;11:2853–63.
Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–64.
Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–8.
Kelly S, Greenman C, Cook PR, Papantonis A. Exon skipping is correlated with exon circularization. J Mol Biol. 2015;427:2414–7.
Zhang XO, Wang HB, Zhang Y, Lu XH, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–47.
Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–34.
Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen LL, et al. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol Cell. 2017;68:940–54 e3.
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66.
Kramer MC, Liang DM, Tatomer DC, Gold B, March ZM, Cherry S, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Gene Dev. 2015;29:2168–82.
Verheijen BM, Pasterkamp RJ. Commentary: FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Front Mol Neurosci. 2017;10:412.
Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3’ processing by human cleavage factor Im. Mol Cell. 2003;12:1467–76.
Yang Q, Coseno M, Gilmartin GM, Doublie S. Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure. 2011;19:368–77.
Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3’-UTRs. Nucleic Acids Res. 2006;34:6264–71.
Tan S, Li H, Zhang W, Shao Y, Liu Y, Guan H, et al. NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene. 2018;37:4887–900.
Sun M, Ding J, Li D, Yang G, Cheng Z, Zhu Q. NUDT21 regulates 3’-UTR length and microRNA-mediated gene silencing in hepatocellular carcinoma. Cancer Lett. 2017;410:158–68.
Park HJ, Ji P, Kim S, Xia Z, Rodriguez B, Li L, et al. 3’ UTR shortening represses tumor-suppressor genes in trans by disrupting ceRNA crosstalk. Nat Genet. 2018;50:783–9.
Lin Y, Li Z, Ozsolak F, Kim SW, Arango-Argoty G, Liu TT, et al. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res. 2012;40:8460–71.
Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3’-UTR landscape across seven tumour types. Nat Commun. 2014;5:5274.
Zhong YX, Du YJ, Yang X, Mo YZ, Fan CM, Xiong F, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79.
Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–16.
Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214–27.
Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–9.
Lin X, Chen Y. Identification of potentially functional CircRNA-miRNA-mRNA regulatory network in hepatocellular carcinoma by integrated microarray analysis. Med Sci Monit Basic Res. 2018;24:70–8.
Gao B, Gao KJ, Li L, Huang ZC, Lin L. miR-184 functions as an oncogenic regulator in hepatocellular carcinoma (HCC). Biomed Pharmacother. 2014;68:143–8.
Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–65.
Yao R, Zou H, Liao W. Prospect of circular RNA in hepatocellular carcinoma: a novel potential biomarker and therapeutic target. Front Oncol. 2018;8:332.
Li P, Sheng C, Huang L, Zhang H, Huang L, Cheng Z, et al. MiR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res. 2014;16:473.
Acknowledgements
We thank the KangChen Bio-tech (Shanghai, China) for providing RNA sequencing and data analysis and the Molecular Medical Center in the Xiangya Hospital (Changsha, China) for the confocal microscopy. We thank Shifu Luo for data analysis. We also thank Dr Wang for kindly supplying the IRES-drived pCircGFP reporter. This research is supported by the National Natural Science Foundation of China (C0709-31201056), the Hunan Provincial Natural Science Foundation of China (2018JJ2493) and the Hunan Provincial Innovation Foundation for Postgraduate (CX20190244).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, X., Ding, J., Wang, X. et al. NUDT21 regulates circRNA cyclization and ceRNA crosstalk in hepatocellular carcinoma. Oncogene 39, 891–904 (2020). https://doi.org/10.1038/s41388-019-1030-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-1030-0
This article is cited by
-
Circular RNAs and their roles in idiopathic pulmonary fibrosis
Respiratory Research (2024)
-
Early-stage idiopathic Parkinson’s disease is associated with reduced circular RNA expression
npj Parkinson's Disease (2024)
-
Exosomal circZNF800 Derived from Glioma Stem-like Cells Regulates Glioblastoma Tumorigenicity via the PIEZO1/Akt Axis
Molecular Neurobiology (2024)
-
The role of circular RNA during the urological cancer metastasis: exploring regulatory mechanisms and potential therapeutic targets
Cancer and Metastasis Reviews (2024)
-
Dihydroartemisinin inhibits restenosis after balloon angioplasty via circHSPA4/miR-19a-5p axis
Molecular and Cellular Biochemistry (2024)