Abstract
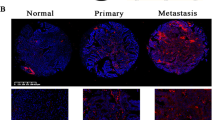
Treatment of ovarian cancer (OvCa) remains challenging owing to its high recurrence rates. Detachment of cancer cells into the peritoneal fluid plays a key role in OvCa relapse, but how this occurs remains incompletely understood. Here we examined global miRNA expression profiles of paired primary/recurrent OvCa specimens and identified a novel biomarker, microRNA-150-5p (miR-150-5p), that was significantly upregulated in 16 recurrent OvCa tissues compared with their matched primary specimens. Analyses of cohorts from two other groups confirmed that expression of miR-150-5p was associated with early relapse and poor survival of OvCa patients. Inhibition of miR-150-5p significantly inhibited the migration and invasion of OvCa cells and induced a mesenchymal-epithelial transition (MET) phenotype. We demonstrated that the proto-oncogene, MYB, is an miR-150-5p target in OvCa cells and that the miR-150-5p/c-Myb/Slug axis plays important roles in regulating epithelial-mesenchymal transition (EMT) in OvCa cells. Expression of MYB was significantly correlated with good clinical outcome in OvCa and was negatively correlated with Slug expression in late-stage clinical specimens. These results suggest that miR-150-5p upregulation mediates the progression of recurrent OvCa by targeting the c-Myb/Slug pathway. Inhibition of miR-150-5p may serve as a new therapeutic strategy for preventing recurrence of OvCa.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Microarray data have been deposited in GEO database (accession number: GSE135469).
References
Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. 2014;384:1376–88.
Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284–96.
Bureau of Health Promotion DoH. The Executive Yuan. Cancer registry annual report, 2016. Taiwan: Bureau of Health Promotion DoH, The Executive Yuan; 2018.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30.
Naora H, Montell DJ. Ovarian cancer metastasis: integrating insights from disparate model organisms. Nat Rev Cancer. 2005;5:355–66.
Smith BN, Bhowmick NA. Role of EMT in metastasis and therapy resistance. J Clin Med. 2016;5:E17.
Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–54.
Bast RC Jr, Klug TL, St John E, Jenison E, Niloff JM, Lazarus H, et al. A radioimmunoassay using a monoclonal antibody to monitor the course of epithelial ovarian cancer. N Engl J Med. 1983;309:883–7.
Mainguene C, Aillet G, Kremer M, Chatal JF. Immunohistochemical study of ovarian tumors using the OC 125 monoclonal antibody as a basis for potential in vivo and in vitro applications. J Nucl Med Allied Sci. 1986;30:19–22.
Goonewardene TI, Hall MR, Rustin GJ. Management of asymptomatic patients on follow-up for ovarian cancer with rising CA-125 concentrations. Lancet Oncol. 2007;8:813–21.
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54.
Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110.
Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004.
Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–55.
Shih KK, Qin LX, Tanner EJ, Zhou Q, Bisogna M, Dao F, et al. A microRNA survival signature (MiSS) for advanced ovarian cancer. Gynecol Oncol. 2011;121:444–50.
Bagnoli M, De Cecco L, Granata A, Nicoletti R, Marchesi E, Alberti P, et al. Identification of a chrXq27.3 microRNA cluster associated with early relapse in advanced stage ovarian cancer patients. Oncotarget. 2011;2:1265–78.
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006.
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8.
Bagnoli M, Canevari S, Califano D, Losito S, Maio MD, Raspagliesi F, et al. Development and validation of a microRNA-based signature (MiROvaR) to predict early relapse or progression of epithelial ovarian cancer: a cohort study. Lancet Oncol. 2016;17:1137–46.
Smolle E, Taucher V, Haybaeck J. Malignant ascites in ovarian cancer and the role of targeted therapeutics. Anticancer Res. 2014;34:1553–61.
Wang F, Zhao X, Tan W, Liu W, Jin Y, Liu Q. Early identification of recurrence in ovarian cancer: a comparison between the ovarian cancer metastasis index and CA-125 levels. PeerJ. 2018;6:e5912.
Vergara D, Merlot B, Lucot JP, Collinet P, Vinatier D, Fournier I, et al. Epithelial-mesenchymal transition in ovarian cancer. Cancer Lett. 2010;291:59–66.
Pradeep S, Kim SW, Wu SY, Nishimura M, Chaluvally-Raghavan P, Miyake T, et al. Hematogenous metastasis of ovarian cancer: rethinking mode of spread. Cancer Cell. 2014;26:77–91.
Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010;177:1053–64.
Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Investig. 2013;123:517–25.
Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–208.
Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15.
Cordero AB, Kwon Y, Hua X, Godwin AK. In vivo imaging and therapeutic treatments in an orthotopic mouse model of ovarian cancer. J Vis Exp. 2010;42:2125.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73.
Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–44.
Xu L, Zhang Y, Qu X, Che X, Guo T, Cai Y, et al. E3 ubiquitin ligase Cbl-b prevents tumor metastasis by maintaining the epithelial phenotype in multiple drug-resistant gastric and breast cancer cells. Neoplasia. 2017;19:374–82.
Gunaje JJ, Bahrami AJ, Schwartz SM, Daum G, Mahoney WM Jr. PDGF-dependent regulation of regulator of G protein signaling-5 expression and vascular smooth muscle cell functionality. Am J Physiol Cell Physiol. 2011;301:C478–89.
Lin YC, Kuo MW, Yu J, Kuo HH, Lin RJ, Lo WL, et al. c-Myb is an evolutionary conserved miR-150 target and miR-150/c-Myb interaction is important for embryonic development. Mol Biol Evol. 2008;25:2189–98.
Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–59.
Kurrey NK, K A, Bapat SA. Snail and Slug are major determinants of ovarian cancer invasiveness at the transcription level. Gynecol Oncol. 2005;97:155–65.
Theriault BL, Shepherd TG, Mujoomdar ML, Nachtigal MW. BMP4 induces EMT and Rho GTPase activation in human ovarian cancer cells. Carcinogenesis. 2007;28:1153–62.
Wang Y, Sheng Q, Spillman MA, Behbakht K, Gu H. Gab2 regulates the migratory behaviors and E-cadherin expression via activation of the PI3K pathway in ovarian cancer cells. Oncogene. 2012;31:2512–20.
Davidson B, Trope CG, Reich R. Epithelial-mesenchymal transition in ovarian carcinoma. Front Oncol. 2012;2:33.
Zhao L, Wang W, Xu L, Yi T, Zhao X, Wei Y, et al. Integrative network biology analysis identifies miR-508-3p as the determinant for the mesenchymal identity and a strong prognostic biomarker of ovarian cancer. Oncogene. 2018;38:2305–19.
Mateescu B, Batista L, Cardon M, Gruosso T, de Feraudy Y, Mariani O, et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17:1627–35.
Konecny GE, Wang C, Hamidi H, Winterhoff B, Kalli KR, Dering J, et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J Natl Cancer Inst. 2014;106:dju249.
Karlan BY, Dering J, Walsh C, Orsulic S, Lester J, Anderson LA, et al. POSTN/TGFBI-associated stromal signature predicts poor prognosis in serous epithelial ovarian cancer. Gynecol Oncol. 2014;132:334–42.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123:725–31.
Cormio G, Rossi C, Cazzolla A, Resta L, Loverro G, Greco P, et al. Distant metastases in ovarian carcinoma. Int J Gynecol Cancer. 2003;13:125–9.
Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV, et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell. 2013;23:186–99.
Sun Y, Hu L, Zheng H, Bagnoli M, Guo Y, Rupaimoole R, et al. MiR-506 inhibits multiple targets in the epithelial-to-mesenchymal transition network and is associated with good prognosis in epithelial ovarian cancer. J Pathol. 2015;235:25–36.
Chan CK, Pan Y, Nyberg K, Marra MA, Lim EL, Jones SJ, et al. Tumour-suppressor microRNAs regulate ovarian cancer cell physical properties and invasive behaviour. Open Biol. 2016;6:160275.
Jia L, Liu W, Cao B, Li H, Yin C. MiR-507 inhibits the migration and invasion of human breastcancer cells through Flt-1 suppression. Oncotarget. 2016;7:36743–54.
Dong P, Xiong Y, Yue J, Hanley SJB, Watari H. miR-34a, miR-424 and miR-513 inhibit MMSET expression to repress endometrial cancer cell invasion and sphere formation. Oncotarget. 2018;9:23253–63.
Zhang J, Luo N, Luo Y, Peng Z, Zhang T, Li S. microRNA-150 inhibits human CD133-positive liver cancer stem cells through negative regulation of the transcription factor c-Myb. Int J Oncol. 2012;40:747–56.
Feng J, Yang Y, Zhang P, Wang F, Ma Y, Qin H, et al. miR-150 functions as a tumour suppressor in human colorectal cancer by targeting c-Myb. J Cell Mol Med. 2014;18:2125–34.
Wang F, Ren X, Zhang X. Role of microRNA-150 in solid tumors. Oncol Lett. 2015;10:11–6.
Li H, Ouyang R, Wang Z, Zhou W, Chen H, Jiang Y, et al. MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci Rep. 2016;6:39001.
Zhang HY, Zheng FS, Yang W, Lu JB. The long non-coding RNA MIAT regulates zinc finger E-box binding homeobox 1 expression by sponging miR-150 and promoteing cell invasion in non-small-cell lung cancer. Gene. 2017;633:61–5.
Wang WH, Chen J, Zhao F, Zhang BR, Yu HS, Jin HY, et al. MiR-150-5p suppresses colorectal cancer cell migration and invasion through targeting MUC4. Asian Pac J Cancer Prev. 2014;15:6269–73.
Guo YH, Wang LQ, Li B, Xu H, Yang JH, Zheng LS, et al. Wnt/beta-catenin pathway transactivates microRNA-150 that promotes EMT of colorectal cancer cells by suppressing CREB signaling. Oncotarget. 2016;7:42513–26.
Narod S. Can advanced-stage ovarian cancer be cured? Nat Rev Clin Oncol. 2016;13:255–61.
Vang S, Wu HT, Fischer A, Miller DH, MacLaughlan S, Douglass E, et al. Identification of ovarian cancer metastatic miRNAs. PLoS ONE. 2013;8:e58226.
Noseda A, Berens ME, Piantadosi C, Modest EJ. Neoplastic cell inhibition with new ether lipid analogs. Lipids. 1987;22:878–83.
Godwin AK, Meister A, O’Dwyer PJ, Huang CS, Hamilton TC, Anderson ME. High resistance to cisplatin in human ovarian cancer cell lines is associated with marked increase of glutathione synthesis. Proc Natl Acad Sci USA. 1992;89:3070–4.
Benard J, Da Silva J, De Blois MC, Boyer P, Duvillard P, Chiric E, et al. Characterization of a human ovarian adenocarcinoma line, IGROV1, in tissue culture and in nude mice. Cancer Res. 1985;45:4970–9.
Hsu IL, Chou CY, Wu YY, Wu JE, Liang CH, Tsai YT, et al. Targeting FXYD2 by cardiac glycosides potently blocks tumor growth in ovarian clear cell carcinoma. Oncotarget. 2016;7:62925–38.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–9.
Acknowledgements
We are grateful for providing the services from the RNAi Core Lab, Research Center of Clinical Medicine, National Cheng Kung University Hospital, Taiwan. RNAi reagents were obtained from the National RNAi Core Facility located at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica, Taiwan. This study was supported by grants MOST 107-2314-B-006-068-MY3, MOST 108-2314-B-006-004, MOST 108-2321-B-006-010, and MOST 106-2314-B-006-015-MY2 from the Ministry of Science and Technology, Taiwan.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Tung, CH., Kuo, LW., Huang, MF. et al. MicroRNA-150-5p promotes cell motility by inhibiting c-Myb-mediated Slug suppression and is a prognostic biomarker for recurrent ovarian cancer. Oncogene 39, 862–876 (2020). https://doi.org/10.1038/s41388-019-1025-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-1025-x
This article is cited by
-
Diverse activity of miR-150 in Tumor development: shedding light on the potential mechanisms
Cancer Cell International (2023)
-
MiR-455-5p suppresses PDZK1IP1 to promote the motility of oral squamous cell carcinoma and accelerate clinical cancer invasion by regulating partial epithelial-to-mesenchymal transition
Journal of Experimental & Clinical Cancer Research (2023)
-
Intricate crosstalk between MYB and noncoding RNAs in cancer
Cancer Cell International (2021)
-
Systematic analysis of the relationship between ovarian cancer prognosis and alternative splicing
Journal of Ovarian Research (2021)
-
FoxP3-miR-150-5p/3p suppresses ovarian tumorigenesis via an IGF1R/IRS1 pathway feedback loop
Cell Death & Disease (2021)