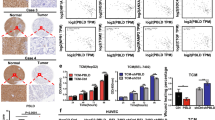
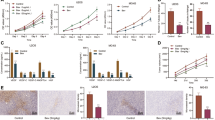
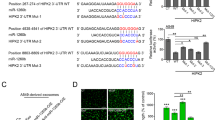
Abstract
Current anti-angiogenic therapy for cancer is based mainly on inhibition of the vascular endothelial growth factor pathway. However, due to the transient and only modest benefit from such therapy, additional approaches are needed. Deregulation of microRNAs (miRNAs) has been demonstrated to be involved in tumor angiogenesis and offers opportunities for a new therapeutic approach. However, effective miRNA-delivery systems are needed for such approaches to be successful. In this study, miRNA profiling of patient data sets, along with in vitro and in vivo experiments, revealed that miR-204-5p could promote angiogenesis in ovarian tumors through THBS1. By binding with scavenger receptor class B type 1 (SCARB1), reconstituted high-density lipoprotein–nanoparticles (rHDL–NPs) were effective in delivering miR-204-5p inhibitor (miR-204-5p-inh) to tumor sites to suppress tumor growth. These results offer a new understanding of miR-204-5p in regulating tumor angiogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–8.
Cao Y. Future options of anti-angiogenic cancer therapy. Chin J Cancer. 2016;35:21.
Al-Abd AM, Alamoudi AJ, Abdel-Naim AB, Neamatallah TA, Ashour OM. Anti-angiogenic agents for the treatment of solid tumors: potential pathways, therapy and current strategies - A review. J Adv Res. 2017;8:591–605.
Niu G, Chen X. Vascular endothelial growth factor as an anti-angiogenic target for cancer therapy. Curr Drug Targets. 2010;11:1000–17.
Dey N, De P, Brian LJ. Evading anti-angiogenic therapy: resistance to anti-angiogenic therapy in solid tumors. Am J Transl Res. 2015;7:1675–98.
Jayson GC, Kerbel R, Ellis LM, Harris AL. Antiangiogenic therapy in oncology: current status and future directions. Lancet. 2016;388:518–29.
Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genom. 2010;11:537–61.
Zhang W, Liu J, Wang G. The role of microRNAs in human breast cancer progression. Tumour Biol. 2014;35:6235–44.
Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, et al. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4:2427.
Wang W, Ren F, Wu Q, Jiang D, Li H, Shi H. MicroRNA-497 suppresses angiogenesis by targeting vascular endothelial growth factor A through the PI3K/AKT and MAPK/ERK pathways in ovarian cancer. Oncol Rep. 2014;32:2127–33.
Li Y, Cai B, Shen L, Dong Y, Lu Q, Sun S, et al. miRNA-29b suppresses tumor growth through simultaneously inhibiting angiogenesis and tumorigenesis by targeting Akt3. Cancer Lett. 2017;397:111–9.
Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions–beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612.
Kuai R, Li D, Chen YE, Moon JJ, Schwendeman A. High-density lipoproteins: nature's multifunctional nanoparticles. ACS Nano. 2016;10:3015–41.
Vickers KC, Remaley AT. HDL and cholesterol: life after the divorce? J Lipid Res. 2014;55:4–12.
Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33.
Mutharasan RK, Foit L, Thaxton CS. High-density lipoproteins for therapeutic delivery systems. J Mater Chem B Mater Biol Med. 2016;4:188–97.
Mooberry LK, Sabnis NA, Panchoo M, Nagarajan B, Lacko AG. Targeting the SR-B1 receptor as a gateway for cancer therapy and imaging. Front Pharmacol. 2016;7:466.
Rajora MA, Zheng G. Targeting SR-BI for cancer diagnostics, imaging and therapy. Front Pharmacol. 2016;7:326.
Lacko AG, Sabnis NA, Nagarajan B, McConathy WJ. HDL as a drug and nucleic acid delivery vehicle. Front Pharmacol. 2015;6:247.
Shahzad MM, Mangala LS, Han HD, Lu C, Bottsford-Miller J, Nishimura M, et al. Targeted delivery of small interfering RNA using reconstituted high-density lipoprotein nanoparticles. Neoplasia. 2011;13:309–19.
Wu SY, Rupaimoole R, Shen F, Pradeep S, Pecot CV, Ivan C, et al. A miR-192-EGR1-HOXB9 regulatory network controls the angiogenic switch in cancer. Nat Commun. 2016;7:11169.
Kuai R, Subramanian C, White PT, Timmermann BN, Moon JJ, Cohen MS, et al. Synthetic high-density lipoprotein nanodisks for targeted withalongolide delivery to adrenocortical carcinoma. Int J Nanomed. 2017;12:6581–94.
Rodrigueza WV, Thuahnai ST, Temel RE, Lund-Katz S, Phillips MC, Williams DL. Mechanism of scavenger receptor class B type I-mediated selective uptake of cholesteryl esters from high density lipoprotein to adrenal cells. J Biol Chem. 1999;274:20344–50.
Inforzato A, Baldock C, Jowitt TA, Holmes DF, Lindstedt R, Marcellini M, et al. The angiogenic inhibitor long pentraxin PTX3 forms an asymmetric octamer with two binding sites for FGF2. J Biol Chem. 2010;285:17681–92.
Lawler J. Thrombospondin-1 as an endogenous inhibitor of angiogenesis and tumor growth. J Cell Mol Med. 2002;6:1–12.
Dunn LL, Simpson PJ, Prosser HC, Lecce L, Yuen GS, Buckle A, et al. A critical role for thioredoxin-interacting protein in diabetes-related impairment of angiogenesis. Diabetes. 2014;63:675–87.
Ohana R, Weiman-Kelman B, Raviv S, Tamm ER, Pasmanik-Chor M, Rinon A, et al. MicroRNAs are essential for differentiation of the retinal pigmented epithelium and maturation of adjacent photoreceptors. Development. 2015;142:2487–98.
Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–64.
Courboulin A, Paulin R, Giguere NJ, Saksouk N, Perreault T, Meloche J, et al. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208:535–48.
Xu G, Chen J, Jing G, Grayson TB, Shalev A. miR-204 targets PERK and regulates UPR Signaling and beta-cell apoptosis. Mol Endocrinol. 2016;30:917–24.
Kaalund SS, Veno MT, Bak M, Moller RS, Laursen H, Madsen F, et al. Aberrant expression of miR-218 and miR-204 in human mesial temporal lobe epilepsy and hippocampal sclerosis-convergence on axonal guidance. Epilepsia. 2014;55:2017–27.
Qiu YH, Wei YP, Shen NJ, Wang ZC, Kan T, Yu WL, et al. miR-204 inhibits epithelial to mesenchymal transition by targeting slug in intrahepatic cholangiocarcinoma cells. Cell Physiol Biochem. 2013;32:1331–41.
Yan H, Wu W, Ge H, Li P, Wang Z. Up-regulation of miR-204 enhances anoikis sensitivity in epithelial ovarian cancer cell line via brain-derived neurotrophic factor pathway in vitro. Int J Gynecol Cancer. 2015;25:944–52.
Wang X, Li F, Zhou X. miR-204-5p regulates cell proliferation and metastasis through inhibiting CXCR4 expression in OSCC. Biomed Pharm. 2016;82:202–7.
Imam JS, Plyler JR, Bansal H, Prajapati S, Bansal S, Rebeles J, et al. Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PLoS One. 2012;7:e52397.
Lee H, Lee S, Bae H, Kang HS, Kim SJ. Genome-wide identification of target genes for miR-204 and miR-211 identifies their proliferation stimulatory role in breast cancer cells. Sci Rep. 2016;6:25287.
Son JC, Jeong HO, Park D, No SG, Lee EK, Lee J, et al. miR-10a and miR-204 as a potential prognostic indicator in low-grade gliomas. Cancer Inform. 2017;16:1176935117702878.
Todorova K, Metodiev MV, Metodieva G, Mincheff M, Fernandez N, Hayrabedyan S. Micro-RNA-204 participates in TMPRSS2/ERG regulation and androgen receptor reprogramming in prostate cancer. Horm Cancer. 2017;8:28–48.
Flores-Perez A, Marchat LA, Rodriguez-Cuevas S, Bautista-Pina V, Hidalgo-Miranda A, Ocampo EA, et al. Dual targeting of ANGPT1 and TGFBR2 genes by miR-204 controls angiogenesis in breast cancer. Sci Rep. 2016;6:34504.
Zhao Y, Zhu CD, Yan B, Zhao JL, Wang ZH. miRNA-directed regulation of VEGF in tilapia under hypoxia condition. Biochem Biophys Res Commun. 2014;454:183–8.
Tatiparti K, Sau S, Kashaw SK, Iyer AK. siRNA delivery strategies: a comprehensive review of recent developments. Nanomaterials (Basel). 2017;7:77.
Yu B, Mao Y, Bai LY, Herman SE, Wang X, Ramanunni A, et al. Targeted nanoparticle delivery overcomes off-target immunostimulatory effects of oligonucleotides and improves therapeutic efficacy in chronic lymphocytic leukemia. Blood. 2013;121:136–47.
Simonsen JB. Evaluation of reconstituted high-density lipoprotein (rHDL) as a drug delivery platform - a detailed survey of rHDL particles ranging from biophysical properties to clinical implications. Nanomedicine. 2016;12:2161–79.
Johnson R, Sabnis N, Sun X, Ahluwalia R, Lacko AG. SR-B1-targeted nanodelivery of anti-cancer agents: a promising new approach to treat triple-negative breast cancer. Breast Cancer (Dove Med Press) 2017;9:383–92.
Zhang F, Wang X, Xu X, Li M, Zhou J, Wang W. Reconstituted high density lipoprotein mediated targeted co-delivery of HZ08 and paclitaxel enhances the efficacy of paclitaxel in multidrug-resistant MCF-7 breast cancer cells. Eur J Pharm Sci. 2016;92:11–21.
Armstrong DK, Bundy B, Wenzel L, Huang HQ, Baergen R, Lele S, et al. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354:34–43.
Lawler PR, Lawler J. Molecular basis for the regulation of angiogenesis by thrombospondin-1 and -2. Cold Spring Harb Perspect Med. 2012;2:a006627.
Resovi A, Pinessi D, Chiorino G, Taraboletti G. Current understanding of the thrombospondin-1 interactome. Matrix Biol. 2014;37:83–91.
Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8.
Zhang J, Guo X, Chang DY, Rosen DG, Mercado-Uribe I, Liu J. CD133 expression associated with poor prognosis in ovarian cancer. Mod Pathol. 2012;25:456–64.
Kang Y, Hu W, Ivan C, Dalton HJ, Miyake T, Pecot CV, et al. Role of focal adhesion kinase in regulating YB-1-mediated paclitaxel resistance in ovarian cancer. J Natl Cancer Inst. 2013;105:1485–95.
Lu C, Han HD, Mangala LS, Ali-Fehmi R, Newton CS, Ozbun L, et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–97.
Acknowledgements
This work is supported, in part, by the National Institutes of Health (CA016672, UH3TR000943, P50 CA217685, P50 CA098258, R35 CA209904), Ovarian Cancer Research Fund, Inc. (Program Project Development Grant), Mr. and Mrs. Daniel P. Gordon, The Blanton-Davis Ovarian Cancer Research Program, the American Cancer Society Research Professor Award, and the Frank McGraw Memorial Chair in Cancer Research (AKS) and CPRIT (DP150091; LSM). SKD was supported by Foundation for Women’s Cancer Grant, CRA was supported by the P50 CA217685 from the SPORE in Ovarian Cancer at MD Anderson.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, X., Mangala, L.S., Mooberry, L. et al. Identifying and targeting angiogenesis-related microRNAs in ovarian cancer. Oncogene 38, 6095–6108 (2019). https://doi.org/10.1038/s41388-019-0862-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0862-y
This article is cited by
-
Association between miR-202, miR-211, and miR-1238 gene polymorphisms and risk of vitiligo
Archives of Dermatological Research (2024)
-
MiR-217 regulates autophagy through OPG/RANKL/RANK in giant cell tumors
Journal of Orthopaedic Surgery and Research (2023)
-
Decreased INPP5B expression predicts poor prognosis in lung adenocarcinoma
Cancer Cell International (2022)
-
Host miRNA and immune cell interactions: relevance in nano-therapeutics for human health
Immunologic Research (2022)
-
Long non-coding RNA MALAT1 enhances angiogenesis during bone regeneration by regulating the miR-494/SP1 axis
Laboratory Investigation (2021)