Abstract
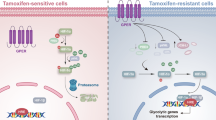
Tamoxifen resistance is one of the major challenges for its medical uses in estrogen receptor (ER)-positive breast cancer. Aerobic glycolysis, an anomalous characteristic of glucose metabolism in cancer cells, has been shown to associate with the resistance to chemotherapeutic agents. It remains, however, largely unclear whether and how tamoxifen resistance contributes to aerobic glycolysis in breast cancer. Here, we report that tamoxifen resistance is associated with enhanced glycolysis in ER-positive breast cancer cells. We demonstrate that EREG, an agonist of EGFR, has an important role in enhancing glycolysis via activating EGFR signaling and its downstream glycolytic genes in tamoxifen-resistant breast cancer cells. We further show that EREG is a direct target of miR-186-3p and that downregulation of miR-186-3p by tamoxifen results in EREG upregulation in tamoxifen-resistant breast cancer cells. Importantly, systemic delivery of cholesterol-modified agomiR-186-3p to mice bearing tamoxifen-resistant breast tumors effectively attenuates both tumor growth and [18F]-fluoro-deoxyglucose ([18F]-FDG) uptake. Together, our results reveal a novel molecular mechanism of resistance to hormone therapies in which the miR-186-3p/EREG axis orchestrates tamoxifen resistance and aerobic glycolysis in ER-positive breast cancer, suggesting targeting miR-186-3p as a promising strategy for therapeutic intervention in endocrine-resistant breast tumors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Jaiyesimi IA, Buzdar AU, Decker DA, Hortobagyi GN. Use of tamoxifen for breast cancer: twenty-eight years later. J Clin Oncol. 1995;13:513–29.
Mills JN, Rutkovsky AC, Giordano A. Mechanisms of resistance in estrogen receptor positive breast cancer: overcoming resistance to tamoxifen/aromatase inhibitors. Curr Opin Pharm. 2018;41:59–65.
Ma CX, Sanchez CG, Ellis MJ. Predicting endocrine therapy responsiveness in breast cancer. Oncol (Williston Park). 2009;23:133–42.
Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717.
Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45:1439–45.
Torres-Arzayus MI, Font de Mora J, Yuan J, Vazquez F, Bronson R, Rue M, et al. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6:263–74.
Johnston SR. Enhancing endocrine therapy for hormone receptor-positive advanced breast cancer: cotargeting signaling pathways. J Natl Cancer Inst. 2015;107:djv212.
Santen RJ, Fan P, Zhang Z, Bao Y, Song RX, Yue W. Estrogen signals via an extra-nuclear pathway involving IGF-1R and EGFR in tamoxifen-sensitive and -resistant breast cancer cells. Steroids. 2009;74:586–94.
Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–82.
Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33.
Swietach P, Vaughan-Jones RD, Harris AL. Regulation of tumor pH and the role of carbonic anhydrase 9. Cancer Metastas Rev. 2007;26:299–310.
Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007;109:3812–9.
Bhattacharya B, Mohd Omar MF, Soong R. The Warburg effect and drug resistance. Br J Pharm. 2016;173:970–9.
Morandi A, Indraccolo S. Linking metabolic reprogramming to therapy resistance in cancer. Biochim Biophys Acta. 2017;1868:1–6.
Knowlden JM, Hutcheson IR, Jones HE, Madden T, Gee JM, Harper ME, et al. Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology. 2003;144:1032–44.
Shelly M, Pinkas-Kramarski R, Guarino BC, Waterman H, Wang LM, Lyass L, et al. Epiregulin is a potent pan-ErbB ligand that preferentially activates heterodimeric receptor complexes. J Biol Chem. 1998;273:10496–505.
Sunaga N, Kaira K, Imai H, Shimizu K, Nakano T, Shames DS, et al. Oncogenic KRAS-induced epiregulin overexpression contributes to aggressive phenotype and is a promising therapeutic target in non-small-cell lung cancer. Oncogene. 2013;32:4034–42.
Qu X, Sandmann T, Frierson H Jr., Fu L, Fuentes E, et al. Integrated genomic analysis of colorectal cancer progression reveals activation of EGFR through demethylation of the EREG promoter. Oncogene. 2016;35:6403–15.
Zhu Z, Kleeff J, Friess H, Wang L, Zimmermann A, Yarden Y, et al. Epiregulin is up-regulated in pancreatic cancer and stimulates pancreatic cancer cell growth. Biochem Biophys Res Commun. 2000;273:1019–24.
Torring N, Jorgensen PE, Sorensen BS, Nexo E. Increased expression of heparin binding EGF (HB-EGF), amphiregulin, TGF alpha and epiregulin in androgen-independent prostate cancer cell lines. Anticancer Res. 2000;20:91–95.
Revillion F, Lhotellier V, Hornez L, Bonneterre J, Peyrat JP. ErbB/HER ligands in human breast cancer, and relationships with their receptors, the bio-pathological features and prognosis. Ann Oncol. 2008;19:73–80.
Zhao S, Liu MF. Mechanisms of microRNA-mediated gene regulation. Sci China C Life Sci. 2009;52:1111–6.
Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23.
Zhang LF, Jiang S, Liu MF. MicroRNA regulation and analytical methods in cancer cell metabolism. Cell Mol Life Sci. 2017;74:2929–41.
Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Ther. 2010;17:523–31.
Thrane S, Pedersen AM, Thomsen MB, Kirkegaard T, Rasmussen BB, Duun-Henriksen AK, et al. A kinase inhibitor screen identifies Mcl-1 and Aurora kinase A as novel treatment targets in antiestrogen-resistant breast cancer cells. Oncogene. 2015;34:4199–210.
Schneider MR, Wolf E. The epidermal growth factor receptor ligands at a glance. J Cell Physiol. 2009;218:460–6.
Lim SO, Li CW, Xia W, Lee HH, Chang SS, Shen J, et al. EGFR signaling enhances aerobic glycolysis in triple-negative breast cancer cells to promote tumor growth and immune escape. Cancer Res. 2016;76:1284–96.
Poliakova M, Aebersold DM, Zimmer Y, Medova M. The relevance of tyrosine kinase inhibitors for global metabolic pathways in cancer. Mol Cancer. 2018;17:27.
Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–33.
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20.
Connor CE, Norris JD, Broadwater G, Willson TM, Gottardis MM, Dewhirst MW, et al. Circumventing tamoxifen resistance in breast cancers using antiestrogens that induce unique conformational changes in the estrogen receptor. Cancer Res. 2001;61:2917–22.
Gee JM, Harper ME, Hutcheson IR, Madden TA, Barrow D, Knowlden JM, et al. The antiepidermal growth factor receptor agent gefitinib (ZD1839/Iressa) improves antihormone response and prevents development of resistance in breast cancer in vitro. Endocrinology. 2003;144:5105–17.
Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S, et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68:826–33.
Peifer C, Alessi DR. New anti-cancer role for PDK1 inhibitors: preventing resistance to tamoxifen. Biochem J. 2009;417:e5–7.
Iorns E, Lord CJ, Ashworth A. Parallel RNAi and compound screens identify the PDK1 pathway as a target for tamoxifen sensitization. Biochem J. 2009;417:361–70.
Daurio NA, Tuttle SW, Worth AJ, Song EY, Davis JM, Snyder NW, et al. AMPK activation and metabolic reprogramming by tamoxifen through estrogen receptor-independent mechanisms suggests new uses for this therapeutic modality in cancer treatment. Cancer Res. 2016;76:3295–306.
Zhu X, Shen H, Yin X, Long L, Xie C, Liu Y, et al. miR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene. 2016;35:323–32.
Ye J, Zhang Z, Sun L, Fang Y, Xu X, Zhou G. miR-186 regulates chemo-sensitivity to paclitaxel via targeting MAPT in non-small cell lung cancer (NSCLC). Mol Biosyst. 2016;12:3417–24.
Li C, Gao Y, Li Y, Ding D. TUG1 mediates methotrexate resistance in colorectal cancer via miR-186/CPEB2 axis. Biochem Biophys Res Commun. 2017;491:552–7.
Sun P, Hu JW, Xiong WJ, Mi J. miR-186 regulates glycolysis through Glut1 during the formation of cancer-associated fibroblasts. Asian Pac J Cancer Prev. 2014;15:4245–50.
Liu L, Wang Y, Bai R, Yang K, Tian Z. MiR-186 inhibited aerobic glycolysis in gastric cancer via HIF-1alpha regulation. Oncogenesis. 2016;5:e224.
Park WC, Liu H, Macgregor Schafer J, Jordan VC. Deregulation of estrogen induced telomerase activity in tamoxifen-resistant breast cancer cells. Int J Oncol. 2005;27:1459–66.
ANSI/ATCC ASN-0002-2011. Authentication of human cell lines: standardization of STR profiling. http://webstore.ansi.org/RecordDetail.aspx?sku=ANSI%2fATCC+ASN-0002-2011.
Wang L, Zhang LF, Wu J, Xu SJ, Xu YY, Li D, et al. IL-1beta-mediated repression of microRNA-101 is crucial for inflammation-promoted lung tumorigenesis. Cancer Res. 2014;74:4720–30.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78.
Zhang LF, Lou JT, Lu MH, Gao C, Zhao S, Li B, et al. Suppression of miR-199a maturation by HuR is crucial for hypoxia-induced glycolytic switch in hepatocellular carcinoma. Embo J. 2015;34:2671–85.
Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH, Liang S, et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. Embo J. 2012;31:1985–98.
Miao Y, Zhang LF, Guo R, Liang S, Zhang M, Shi S, et al. (18)F-FDG PET/CT for monitoring the response of breast cancer to miR-143-based therapeutics by targeting tumor glycolysis. Mol Ther Nucleic Acids. 2016;5:e357.
Gambhir SS, Bauer E, Black ME, Liang Q, Kokoris MS, Barrio JR, et al. A mutant herpes simplex virus type 1 thymidine kinase reporter gene shows improved sensitivity for imaging reporter gene expression with positron emission tomography. Proc Natl Acad Sci USA. 2000;97:2785–90.
Acknowledgements
We thank members of the Chen’s and Zhu’s labs for helpful comments. This work was supported by grants from the National Natural Science Foundation of China (81272544 and 81502267) and Foundation and frontier science and technology project of Yuzhong district science and technology commission, Chongqing, China (20150121).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
He, M., Jin, Q., Chen, C. et al. The miR-186-3p/EREG axis orchestrates tamoxifen resistance and aerobic glycolysis in breast cancer cells. Oncogene 38, 5551–5565 (2019). https://doi.org/10.1038/s41388-019-0817-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0817-3
This article is cited by
-
Metabolic adaptation towards glycolysis supports resistance to neoadjuvant chemotherapy in early triple negative breast cancers
Breast Cancer Research (2024)
-
Metabolism-regulating non-coding RNAs in breast cancer: roles, mechanisms and clinical applications
Journal of Biomedical Science (2024)
-
GPER-mediated stabilization of HIF-1α contributes to upregulated aerobic glycolysis in tamoxifen-resistant cells
Oncogene (2023)
-
KIF2C accelerates the development of non-small cell lung cancer and is suppressed by miR-186-3p via the AKT-GSK3β-β-catenin pathway
Scientific Reports (2023)
-
Non-coding RNAs in breast cancer: with a focus on glucose metabolism reprogramming
Discover Oncology (2023)