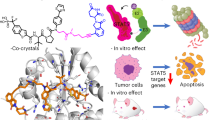
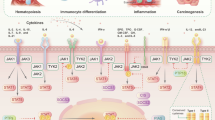
Abstract
The signal transducer and activator of transcription (STAT) are transcription factors that work via JAK/STAT pathway regulating the expression of genes involved in cell survival, proliferation, differentiation, development, immune response, and, among other essential biological functions, hematopoiesis. JAK/STAT signaling is strictly regulated under normal physiological conditions. However, a large group of diverse diseases has been associated to an aberrant regulation of STAT factors. Erroneous modulation of the pathway leads to constitutive STAT activation, thereby driving proliferation, inflammation, and an uncontrolled immune response. Deregulated STAT5 activation has been found in the development of many hematopoietic tumors, including chronic and acute leukemias, polycythemia vera, and lymphoma. Mutations in the kinases that phosphorylate STAT5, and/or overexpression of the upstream receptor-associated tyrosine kinases have been suggested as the main drivers of constitutive STAT5 activation. Hyper-activated STAT5 leads to the aberrant expression of its target genes including antiapoptotic, proliferative, and pro-inflammatory genes, favouring tumorigenesis. In this review, we intent to discuss the biology of JAK/STAT pathway, with particular focus on STAT5 and its crucial role in the development and progression of hematologic malignancies. Furthermore, we provide a synopsis of potential therapeutic strategies based on STAT5 activity inhibition that may represent an excellent opportunity for drug development in oncohematology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
O’Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–28.
Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117(Pt 8):1281–3.
Kiu H, Nicholson SE. Biology and significance of the JAK/STAT signalling pathways. Growth Factors. 2012;30:88–106.
Miklossy G, Hilliard TS, Turkson J. Therapeutic modulators of STAT signalling for human diseases. Nat Rev Drug Discov. 2013;12:611–29.
Sonkin D, Palmer M, Rong X, Horrigan K, Regnier CH, Fanton C, et al. The identification and characterization of a STAT5 gene signature in hematologic malignancies. Cancer Biomark. 2015;15:79–87.
Bibi S, Arslanhan MD, Langenfeld F, Jeanningros S, Cerny-Reiterer S, Hadzijusufovic E, et al. Co-operating STAT5 and AKT signaling pathways in chronic myeloid leukemia and mastocytosis: possible new targets of therapy. Haematologica. 2014;99:417–29.
Ghoreschi K, Laurence A, O’Shea JJ. Janus kinases in immune cell signaling. Immunol Rev. 2009;228:273–87.
Vainchenker W, Dusa A, Constantinescu SN. JAKs in pathology: role of Janus kinases in hematopoietic malignancies and immunodeficiencies. Semin Cell Dev Biol. 2008;19:385–93.
Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, et al. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem. 2001;276:6675–88.
Schindler C, Plumlee C. Inteferons pen the JAK-STAT pathway. Semin Cell Dev Biol. 2008;19:311–8.
Haura EB, Turkson J, Jove R. Mechanisms of disease: insights into the emerging role of signal transducers and activators of transcription in cancer. Nat Clin Pract Oncol. 2005;2:315–24.
Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. J Biol Chem. 2003;278:34133–40.
Yang S, Park K, Turkson J, Arteaga CL. Ligand-independent phosphorylation of Y869 (Y845) links mutant EGFR signaling to stat-mediated gene expression. Exp Cell Res. 2008;314:413–9.
Hu X, Dutta P, Tsurumi A, Li J, Wang J, Land H, et al. Unphosphorylated STAT5A stabilizes heterochromatin and suppresses tumor growth. Proc Natl Acad Sci USA. 2013;110:10213–8.
Fernández-Pérez L, Flores-Morales A, Guerra B, Díaz-Chico JC, Iglesias-Gato D. Growth hormone receptor signaling pathways and its negative regulation by SOCS2. In: Aguayo MdCC-, editor. Restricted growth: clinical, genetic and molecular aspects. InTechOpen; 2016. p. 125–45.
Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–11.
O’Sullivan LA, Liongue C, Lewis RS, Stephenson SE, Ward AC. Cytokine receptor signaling through the Jak-Stat-Socs pathway in disease. Mol Immunol. 2007;44:2497–506.
Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1994;13:2182–91.
Able AA, Burrell JA, Stephens JM. STAT5-interacting proteins: a synopsis of proteins that regulate Stat5 activity. Biology (Basel). 2017;6:pii: E20.
Hughes K, Watson CJ. The spectrum of STAT functions in mammary gland development. JAKSTAT. 2012;1:151–8.
Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282:20059–63.
Wiwi CA, Waxman DJ. Role of hepatocyte nuclear factors in growth hormone-regulated, sexually dimorphic expression of liver cytochromes P450. Growth Factors. 2004;22:79–88.
Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol. 2007;178:2623–9.
Wierenga AT, Vellenga E, Schuringa JJ. Maximal STAT5-induced proliferation and self-renewal at intermediate STAT5 activity levels. Mol Cell Biol. 2008;28:6668–80.
Heltemes-Harris LM, Farrar MA. The role of STAT5 in lymphocyte development and transformation. Curr Opin Immunol. 2012;24:146–52.
Park JH, Adoro S, Guinter T, Erman B, Alag AS, Catalfamo M, et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol. 2010;11:257–64.
Hoelbl A, Kovacic B, Kerenyi MA, Simma O, Warsch W, Cui Y, et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–906.
Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–75.
Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2010;11:171–9.
Kemp RA, Pearson CF, Cornish GH, Seddon BP. Evidence of STAT5-dependent and -independent routes to CD8 memory formation and a preferential role for IL-7 over IL-15 in STAT5 activation. Immunol Cell Biol. 2010;88:213–9.
Wang Z, Bunting KD. STAT5 in hematopoietic stem cell biology and transplantation. JAKSTAT. 2013;2:e27159.
Xiao W, Hong H, Kawakami Y, Lowell CA, Kawakami T. Regulation of myeloproliferation and M2 macrophage programming in mice by Lyn/Hck, SHIP, and Stat5. J Clin Invest. 2008;118:924–34.
Fievez L, Desmet C, Henry E, Pajak B, Hegenbarth S, Garze V, et al. STAT5 is an ambivalent regulator of neutrophil homeostasis. PLoS One. 2007;2:e727.
Dolznig H, Grebien F, Deiner EM, Stangl K, Kolbus A, Habermann B, et al. Erythroid progenitor renewal versus differentiation: genetic evidence for cell autonomous, essential functions of EpoR, Stat5 and the GR. Oncogene. 2006;25:2890–900.
Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. Ineffective erythropoiesis in Stat5a(-/-)5b(-/-) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–73.
Kerenyi MA, Grebien F, Gehart H, Schifrer M, Artaker M, Kovacic B, et al. Stat5 regulates cellular iron uptake of erythroid cells via IRP-2 and TfR-1. Blood. 2008;112:3878–88.
Wang X, Huang S, Chen JL. Understanding of leukemic stem cells and their clinical implications. Mol Cancer. 2017;16:2.
Schepers H, van Gosliga D, Wierenga AT, Eggen BJ, Schuringa JJ, Vellenga E. STAT5 is required for long-term maintenance of normal and leukemic human stem/progenitor cells. Blood. 2007;110:2880–8.
Linher-Melville K, Singh G. The complex roles of STAT3 and STAT5 in maintaining redox balance: Lessons from STAT-mediated xCT expression in cancer cells. Mol Cell Endocrinol. 2017;451:40–52.
Cumaraswamy AA, Lewis AM, Geletu M, Todic A, Diaz DB, Cheng XR, et al. Nanomolar-potency small molecule inhibitor of STAT5 protein. ACS Med Chem Lett. 2014;5:1202–6.
Van Etten RA. Aberrant cytokine signaling in leukemia. Oncogene. 2007;26:6738–49.
Nelson EA, Walker SR, Weisberg E, Bar-Natan M, Barrett R, Gashin LB, et al. The STAT5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors. Blood. 2011;117:3421–9.
Melo JV, Deininger MW. Biology of chronic myelogenous leukemia—signaling pathways of initiation and transformation. Hematol Oncol Clin North Am. 2004;18:545–68.
Warsch W, Grundschober E, Sexl V. Adding a new facet to STAT5 in CML: multitasking for leukemic cells. Cell Cycle. 2013;12:1813–4.
Warsch W, Kollmann K, Eckelhart E, Fajmann S, Cerny-Reiterer S, Holbl A, et al. High STAT5 levels mediate imatinib resistance and indicate disease progression in chronic myeloid leukemia. Blood. 2011;117:3409–20.
Warsch W, Grundschober E, Berger A, Gille L, Cerny-Reiterer S, Tigan AS, et al. STAT5 triggers BCR-ABL1 mutation by mediating ROS production in chronic myeloid leukaemia. Oncotarget. 2012;3:1669–87.
Chatain N, Ziegler P, Fahrenkamp D, Jost E, Moriggl R, Schmitz-Van de Leur H, et al. Src family kinases mediate cytoplasmic retention of activated STAT5 in BCR-ABL-positive cells. Oncogene. 2013;32:3587–97.
Kaymaz BT, Selvi N, Gokbulut AA, Aktan C, Gunduz C, Saydam G, et al. Suppression of STAT5A and STAT5B chronic myeloid leukemia cells via siRNA and antisense-oligonucleotide applications with the induction of apoptosis. Am J Blood Res. 2013;3:58–70.
Kosova B, Tezcanli B, Ekiz HA, Cakir Z, Selvi N, Dalmizrak A, et al. Suppression of STAT5A increases chemotherapeutic sensitivity in imatinib-resistant and imatinib-sensitive K562 cells. Leuk Lymphoma. 2010;51:1895–901.
Zhao R, Xing S, Li Z, Fu X, Li Q, Krantz SB, et al. Identification of an acquired JAK2 mutation in polycythemia vera. J Biol Chem. 2005;280:22788–92.
Spivak JL. Polycythemia vera. Curr Treat Options Oncol. 2018;19:12.
Trung LQ, Espinoza JL, An DT, Viet NH, Shimoda K, Nakao S. Resveratrol selectively induces apoptosis in malignant cells with the JAK2V617F mutation by inhibiting the JAK2 pathway. Mol Nutr Food Res. 2015;59:2143–54.
Yan D, Hutchison RE, Mohi G. Critical requirement for Stat5 in a mouse model of polycythemia vera. Blood. 2012;119:3539–49.
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21.
Hospital MA, Green AS, Maciel TT, Moura IC, Leung AY, Bouscary D, et al. FLT3 inhibitors: clinical potential in acute myeloid leukemia. Onco Targets Ther. 2017;10:607–15.
Dorritie KA, McCubrey JA, Johnson DE. STAT transcription factors in hematopoiesis and leukemogenesis: opportunities for therapeutic intervention. Leukemia. 2014;28:248–57.
Walters DK, Mercher T, Gu TL, O’Hare T, Tyner JW, Loriaux M, et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell. 2006;10:65–75.
Quentmeier H, MacLeod RA, Zaborski M, Drexler HG. JAK2 V617F tyrosine kinase mutation in cell lines derived from myeloproliferative disorders. Leukemia. 2006;20:471–6.
Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109:1139–42.
Tsuruyama T, Nakamura T, Jin G, Ozeki M, Yamada Y, Hiai H. Constitutive activation of Stat5a by retrovirus integration in early pre-B lymphomas of SL/Kh strain mice. Proc Natl Acad Sci USA. 2002;99:8253–8.
Joliot V, Cormier F, Medyouf H, Alcalde H, Ghysdael J. Constitutive STAT5 activation specifically cooperates with the loss of p53 function in B-cell lymphomagenesis. Oncogene. 2006;25:4573–84.
Tasian SK, Doral MY, Borowitz MJ, Wood BL, Chen IM, Harvey RC, et al. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood. 2012;120:833–42.
Walz C, Ahmed W, Lazarides K, Betancur M, Patel N, Hennighausen L, et al. Essential role for Stat5a/b in myeloproliferative neoplasms induced by BCR-ABL1 and JAK2(V617F) in mice. Blood. 2012;119:3550–60.
Swaminathan S, Klemm L, Park E, Papaemmanuil E, Ford A, Kweon SM, et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16:766–74.
Lee S, Shah T, Yin C, Hochberg J, Ayello J, Morris E, et al. Ruxolitinib significantly enhances in vitro apoptosis in Hodgkin lymphoma and primary mediastinal B-cell lymphoma and survival in a lymphoma xenograft murine model. Oncotarget. 2018;9:9776–88.
Zahn M, Marienfeld R, Melzner I, Heinrich J, Renner B, Wegener S, et al. A novel PTPN1 splice variant upregulates JAK/STAT activity in classical Hodgkin lymphoma cells. Blood. 2017;129:1480–90.
Jona A, Szodoray P, Illes A. Immunologic pathomechanism of Hodgkin’s lymphoma. Exp Hematol. 2013;41:995–1004.
Mata E, Diaz-Lopez A, Martin-Moreno AM, Sanchez-Beato M, Varela I, Mestre MJ, et al. Analysis of the mutational landscape of classic Hodgkin lymphoma identifies disease heterogeneity and potential therapeutic targets. Oncotarget. 2017;8:111386–95.
Scheeren FA, Diehl SA, Smit LA, Beaumont T, Naspetti M, Bende RJ, et al. IL-21 is expressed in Hodgkin lymphoma and activates STAT5: evidence that activated STAT5 is required for Hodgkin lymphomagenesis. Blood. 2008;111:4706–15.
Scheeren FA, Naspetti M, Diehl S, Schotte R, Nagasawa M, Wijnands E, et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat Immunol. 2005;6:303–13.
Re D, Thomas RK, Behringer K, Diehl V. From Hodgkin disease to Hodgkin lymphoma: biologic insights and therapeutic potential. Blood. 2005;105:4553–60.
Martin-Subero JI, Gesk S, Harder L, Sonoki T, Tucker PW, Schlegelberger B, et al. Recurrent involvement of the REL and BCL11A loci in classical Hodgkin lymphoma. Blood. 2002;99:1474–7.
Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198:851–62.
Kuppers R. Molecular biology of Hodgkin’s lymphoma. Adv Cancer Res. 2002;84:277–312.
Moslehi JJ, Deininger M. Tyrosine kinase inhibitor-associated cardiovascular toxicity in chronic myeloid leukemia. J Clin Oncol. 2015;33:4210–8.
Kayser S, Levis MJ. FLT3 tyrosine kinase inhibitors in acute myeloid leukemia: clinical implications and limitations. Leuk Lymphoma. 2014;55:243–55.
Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–83.
Guerra B, Martin-Rodriguez P, Diaz-Chico JC, McNaughton-Smith G, Jimenez-Alonso S, Hueso-Falcon I, et al. CM363, a novel naphthoquinone derivative which acts as multikinase modulator and overcomes imatinib resistance in chronic myelogenous leukemia. Oncotarget. 2017;8:29679–98.
Szelag M, Wesoly J, Bluyssen HA. Advances in peptidic and peptidomimetic-based approaches to inhibit STAT signaling in human diseases. Curr Protein Pept Sci. 2016;17:135–46.
Wingelhofer B, Maurer B, Heyes EC, Cumaraswamy AA, Berger-Becvar A, de Araujo ED, et al. Pharmacologic inhibition of STAT5 in acute myeloid leukemia. Leukemia. 2018;32:1135–46.
Sun J, McKallip RJ. Plumbagin treatment leads to apoptosis in human K562 leukemia cells through increased ROS and elevated TRAIL receptor expression. Leuk Res. 2011;35:1402–8.
Martín-Rodríguez P, Guerra B, Hueso-Falcon I, Aranda-Tavío H, Diaz-Chico J, Quintana J, et al. A novel naphthoquinone-coumarin hybrid that inhibits BCR-ABL1-STAT5 oncogenic pathway and reduces survival in imatinib-resistant Chronic Myelogenous Leukemia cells. Front Pharmacol 2018;9:1546.
Nelson EA, Walker SR, Xiang M, Weisberg E, Bar-Natan M, Barrett R, et al. The STAT5 inhibitor pimozide displays efficacy in models of acute myelogenous leukemia driven by FLT3 mutations. Genes Cancer. 2012;3:503–11.
Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58.
Tsatsanis C, Spandidos DA. The role of oncogenic kinases in human cancer (Review). Int J Mol Med. 2000;5:583–90.
Campia I, Buondonno I, Castella B, Rolando B, Kopecka J, Gazzano E, et al. An autocrine cytokine/JAK/STAT-signaling induces kynurenine synthesis in multidrug resistant human cancer cells. PLoS One. 2015;10:e0126159.
Hochhaus A, O’Brien SG, Guilhot F, Druker BJ, Branford S, Foroni L, et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23:1054–61.
de Lavallade H, Apperley JF, Khorashad JS, Milojkovic D, Reid AG, Bua M, et al. Imatinib for newly diagnosed patients with chronic myeloid leukemia: incidence of sustained responses in an intention-to-treat analysis. J Clin Oncol. 2008;26:3358–63.
Reich NC. STATs get their move on. JAKSTAT. 2013;2:e27080.
Stark GR, Cheon H, Wang Y. Responses to cytokines and interferons that depend upon JAKs and STATs. Cold Spring Harb Perspect Biol. 2018;10:pii: a028555.
Acknowledgements
We thank all the authors that contributed to the understanding of the role of STAT5 in hematopoiesis and oncohematology. We apologize to those whose work deserves to be cited but unfortunately are not quoted because of space restriction. The research program in the author’s lab was supported by grants-in-aid from Spanish Ministry of Economy and Competitivity (MINECO) with the funding of European Regional Development Fund-European Social Fund (SAF2015-65113-C2-2-R) and Alfredo Martin-Reyes Foundation (Arehucas) - FICIC. CR and MMG were supported by postdoctoral grants-in-aid from MINECO - Juan de la Cierva 2017 program and ULPGC 2017, respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Recio, C., Guerra, B., Guerra-Rodríguez, M. et al. Signal transducer and activator of transcription (STAT)-5: an opportunity for drug development in oncohematology. Oncogene 38, 4657–4668 (2019). https://doi.org/10.1038/s41388-019-0752-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0752-3
This article is cited by
-
JAK2/STAT3 inhibition attenuates intestinal ischemia–reperfusion injury via promoting autophagy: in vitro and in vivo study
Molecular Biology Reports (2022)
-
Roles for macrophage-polarizing interleukins in cancer immunity and immunotherapy
Cellular Oncology (2022)
-
STAT proteins: a kaleidoscope of canonical and non-canonical functions in immunity and cancer
Journal of Hematology & Oncology (2021)