Abstract
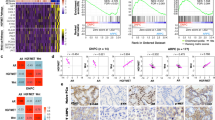
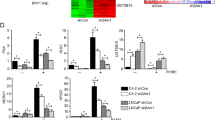
Nuclear receptor tyrosine kinases (nRTKs) are aberrantly upregulated in many types of cancers, but the regulation of nRTK remains unclear. We previously showed androgen deprivation therapy (ADT) induces nMET in castration-resistant prostate cancer (CRPC) specimens. Through gene expression microarray profiles reanalysis, we identified that nMET signaling requires ARF for CRPC growth in Pten/Trp53 conditional knockout mouse model. Accordingly, aberrant MET/nMET elevation correlates with ARF in human prostate cancer (PCa) specimens. Mechanistically, ARF elevates nMET through binding to MET cytoplasmic domain to stabilize MET. Furthermore, carbon nanodots resensitize cancer cells to MET inhibitors through DNA damage response. The inhibition of phosphorylation by carbon nanodots was identified through binding to phosphate group of phospho-tyrosine via computational calculation and experimental assay. Thus, nMET is essential to precision therapy of MET inhibitor. Our findings reveal for the first time that targeting nMET axis by carbon nanodots can be a novel avenue for overcoming drug resistance in cancers especially prostate cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin. 1972;22:232–40.
Chen WY, Tsai YC, Yeh HL, Suau F, Jiang KC, Shao AN, et al. Loss of SPDEF and gain of TGFBI activity after androgen deprivation therapy promote EMTand bone metastasis of prostate cancer. Sci Signal. 2017;10:492.
Petrylak DP, Tangen CM, Hussain MH, Lara PNJ, Jones JA, Taplin ME, et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351:1513–20.
Domingo-Domenech J, Vidal SJ, Rodriguez-Bravo V, Castillo-Martin M, Quinn SA, Rodriguez-Barrueco R, et al. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer Cell. 2012;22:373–88.
Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13.
Xie Y, Xu K, Linn DE, Yang X, Guo Z, Shimelis H, et al. The 44-kDa Pim-1 kinase phosphorylates BCRP/ABCG2 and thereby promotes its multimerization and drug-resistant activity in human prostate cancer cells. J Biol Chem. 2008;283:3349–56.
Xie Y, Burcu M, Linn DE, Qiu Y, Baer MR. Pim-1 kinase protects P-glycoprotein from degradation and enables its glycosylation and cell surface expression. Mol Pharmacol. 2010;78:310–8.
Guo Z, Dai B, Jiang T, Xu K, Xie Y, Kim O, et al. Regulation of androgen receptor activity by tyrosine phosphorylation. Cancer Cell. 2006;10:309–19.
Verras M, Lee J, Xue H, Li TH, Wang Y, Sun Z. The androgen receptor negatively regulates the expression of c-Met: implications for a novel mechanism of prostate cancer progression. Cancer Res. 2007;67:967–75.
Gherardi E, Birchmeier W, Birchmeier C, Vande WG. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103.
Wanjala J, Taylor BS, Chapinski C, Hieronymus H, Wongvipat J, Chen Y, et al. Identifying actionable targets through integrative analyses of GEM model and human prostate cancer genomic profiling. Mol Cancer Ther. 2015;14:278–88.
Pozner-Moulis S, Pappas DJ, Rimm DL. Met, the hepatocyte growth factor receptor, localizes to the nucleus in cells at low density. Cancer Res. 2006;66:7976–82.
Xie Y, Lu W, Liu S, Yang Q, Carver BS, Li E, et al. Crosstalk between nuclear MET and SOX9/beta-catenin correlates with castration-resistant prostate cancer. Mol Endocrinol. 2014;28:1629–39.
Tu WH, Zhu C, Clark C, Christensen JG, Sun Z. Efficacy of c-Met inhibitor for advanced prostate cancer. BMC Cancer. 2010;10:556.
Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med. 2012;4:120ra117.
Song A, Kim TM, Kim DW, Kim S, Keam B, Lee SH, et al. Molecular changes associated with acquired resistance to crizotinib in ROS1-rearranged non-small cell lung cancer (NSCLC). Clin Cancer Res. 2015;21:2379–87.
Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–75.
Chen Z, Carracedo A, Lin HK, Koutcher JA, Behrendt N, Egia A, et al. Differential p53-independent outcomes ofp19(Arf) loss in oncogenesis. Sci Signal. 2009;2:ra44.
Xie Y, Liu S, Lu W, Yang Q, Williams KD, Binhazim AA, et al. Slug regulates E-cadherin repression via p19Arf in prostate tumorigenesis. Mol Oncol. 2014;8:1355–64.
Xie Y, Lu W, Liu S, Yang Q, Goodwin JS, Sathyanarayana SA, et al. MMP7 interacts with ARF in nucleus to potentiate tumor microenvironments for prostate cancer progression in vivo. Oncotarget. 2016;7:47609–19.
Vivo M, Fontana R, Ranieri M, Capasso G, Angrisano T, Pollice A, et al. p14ARF interacts with the focal adhesion kinase and protects cells from anoikis. Oncogene. 2017;36:4913–28.
Humbey O, Pimkina J, Zilfou JT, Jarnik M, Dominguez-Brauer C, Burgess DJ, et al. The ARF tumor suppressor can promote the progression of some tumors. Cancer Res. 2008;68:9608–13.
Li CL, Ou CM, Wu WC, Chen YP, Lin TE, Ho LC, et al. Carbon dots prepared from ginger exhibiting efficient inhibition of human hepatocellular carcinoma cells. J Mater Chem B. 2014;2:4564–71.
Xie Y, Filchakova O, Yang Q, Yesbolatov Y, Tursynkhan D, Kassymbek A, et al. Inhibition of cancer cell proliferation by carbon dots derived from date pits at low dose. ChemistrySelect. 2017;2:4079–83.
Xie Y, Sun Q, Nurkesh AA, Lu J, Kauanova S, Feng J, et al. Dysregulation of YAP by ARF stimulated with tea-derived carbon nanodots. Sci Rep. 2017;7:16577.
Ding HM, Ma YQ. Computational approaches to cell-nanomaterial interactions: keeping balance between therapeutic efficiency and cytotoxicity. Nanoscale Horiz. 2018;3:6–27.
Liu S, Meng XY, Perez-Aguilar JM, Zhou R. An in silico study of TiO(2)nanoparticles interaction with twenty standard amino acids in aqueous solution. Sci Rep. 2016;6:37761.
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;16:1781–802.
Likos CN. Effective interactions in soft condensed matter physics. Phys Rep. 2001;348:267–439.
Vácha R, Martinez-Veracoechea FJ, Frenkel D. Receptor-mediated endocytosis of nanoparticles of various shapes. Nano Lett. 2011;11:5391–5.
Kaur D, Khanna S. Intermolecular hydrogen bonding interactions of furan, isoxazole and oxazole with water. Comput Theor Chem. 2011;963:71–75.
Klähn M, Mathias G, Kötting C, Nonella M, Schlitter J, Gerwert K, et al. IR spectra of phosphate ions in aqueous solution: predictions of a DFT/MM approach compared with observations. J Phy Chem A. 2004;108:6186–94.
Mudryj M, Siddiqui S, Libertini SJ, Lombard AP, Mooso B, D’Abronzo L, et al. Androgen receptor-mediated regulation of p14ARF transcription in prostate tumor cells. [abstract]. In: Proceedings of the 106th Annual Meeting of the American Association for Cancer Research; 2015; Philadelphia, PA. Philadelphia (PA): AACR; Cancer Res. 2015;75(15 Suppl): Abstract no. 5051. https://doi.org/10.1158/1538-7445.AM2015-5051.
Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30.
Lachat Y, Diserens AC, Nozaki M, Kobayashi H, Hamou MF, Godard S, et al. INK4a/Arf is required for suppression of EGFR/DeltaEGFR(2-7)-dependent ERK activation in mouse astrocytes and glioma. Oncogene. 2004;23:6854–63.
Yu J, Zhang SS, Saito K, Williams S, Arimura Y, Ma Y, et al. PTEN regulation by Akt-EGR1-ARF-PTEN axis. EMBO. 2009;28:21–33.
Khoo CM, Carrasco DR, Bosenberg MW, Paik JH, Depinho RA. Ink4a/Arf tumor suppressor does not modulate the degenerative conditions or tumor spectrum of the telomerase-deficient mouse. Proc Natl Acad Sci USA. 2007;104:3931–6.
Zhang M, Liu E, Cui Y, Huang Y. Nanotechnology-based combination therapy for overcoming multidrug-resistant cancer. Cancer Biol Med. 2017;14:212–27.
Yuan Y, Guo B, Hao L, Liu N, Lin Y, Guo W, et al. Doxorubicin-loadedenvironmentally friendly carbon dots as a novel drug delivery system for nucleus targeted cancer therapy. Colloids Surf B Biointerfaces. 2017;159:349–59.
Mizrachi A, Shamay Y, Shah J, Brook S, Soong J, Rajasekhar VK, et al. Tumour-specific PI3K inhibition via nanoparticle-targeted delivery in head and neck squamous cell carcinoma. Nat Commun. 2017;8:14292.
Petrushev B, Boca S, Simon T, Berce C, Frinc I, Dima D, et al. Gold nanoparticles enhance the effect of tyrosine kinase inhibitors in acute myeloid leukemia therapy. Int J Nanomed. 2016;11:641–60.
Xie Y, Istayeva S, Chen Z, Tokay T, Zhumadilov Z, Wu D, et al. nMET, a new target in recurrent cancer. Curr Cancer Drug Targets. 2016;16:572–8.
Kim SM, Kim H, Yun MR, Kang HN, Pyo KH, Park HJ, et al. Activation of the Met kinase confers acquired drug resistance in FGFR-targeted lung cancer therapy. Oncogenesis. 2016;5:e241.
Ahn SY, Kim J, Kim MA, Choi J, Kim WH. Increased HGF expression induces resistance to c-MET tyrosine kinase inhibitors in gastric cancer. Anticancer Res. 2017;37:1127–38.
Lu W, Xie Y, Ma Y, Matusik RJ, Chen Z. ARF represses androgen receptor transactivation in prostate cancer. Mol Endocrinol. 2013;27:635–48.
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57.
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462.
Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30.
Ji H, Wang J, Nika H, Hawke D, Keezer S, Ge Q, et al. EGF-induced ERK activation promotes CK2-mediated disassociation of alpha-Catenin from beta-Catenin and transactivation of beta-Catenin. Mol Cell. 2009;36:547–59.
Acknowledgements
We would like to thank Drs. Simon W. Hayward and LaMonica Stewart for cell lines, Lynn M. Matrisian for providing MMP7-Luc plasmid. This work was supported in part by NIH grants MD004038, MD007586, CA091408 and CA163069, UL1 RR024975-01 and UL1 TR000445-06. IF and IHC analysis was performed by Meharry Medical College Morphology Core, which is supported in part by NIH grants U54 MD007593, G12 MD007586, R24 DA036420, and S10RR0254970. We would like to thank Dr. Guoliang Li, Zhanna Alexeyeva, Aigerim Kabulova, Zhibek Keneskhanova, Akerke Altaikyzy for the kind help or assistant on research. Quantum mechanical calculations were performed on the clusters of the Siberian Supercomputer Center (http://www2.sscc.ru) of the Institute of Computational Mathematics and Mathematical Geophysics of the Siberian Branch of the Russian Academy of Sciences.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Rights and permissions
About this article
Cite this article
Xie, Y., Fan, H., Lu, W. et al. Nuclear MET requires ARF and is inhibited by carbon nanodots through binding to phospho-tyrosine in prostate cancer. Oncogene 38, 2967–2983 (2019). https://doi.org/10.1038/s41388-018-0608-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0608-2
This article is cited by
-
p53 modulates kinase inhibitor resistance and lineage plasticity in NF1-related MPNSTs
Oncogene (2024)
-
Novel carbon nanozymes with enhanced phosphatase-like catalytic activity for antimicrobial applications
Discover Nano (2023)
-
Carbon nanoparticles induce DNA repair and PARP inhibitor resistance associated with nanozyme activity in cancer cells
Cancer Nanotechnology (2022)