Abstract
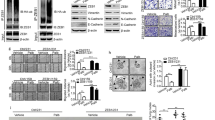
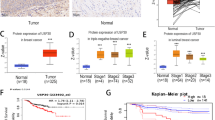
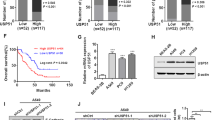
Metastatic progression is the main cause of mortality in breast cancer, necessitating the determination of the molecular events driving this process for the development of new therapeutic approaches. Here, we demonstrate that hyperactivation of the deubiquitinase USP1 contributes to breast cancer metastasis. Upregulated USP1 expression in primary breast cancer specimens correlates with metastatic progression and poor prognosis in breast cancer patients. USP1 enhances the expression of a number of pro-metastatic genes in breast cancer cells, promotes cell migration and invasion in vitro, and facilitates lung metastasis of breast cancer cells. Moreover, USP1-mediated deubiquitination and stabilization of KPNA2 are revealed as the downstream events crucial for USP1-pro-metastatic function. Most importantly, pharmacological intervention of USP1 function by pimozide or ML323 significantly represses breast cancer metastasis in mice, suggesting a rationale for using USP1 inhibitors for treatment of patients with breast cancer. Taken together, our results establish USP1 as a promoter of breast cancer metastasis and provide evidence for the potential practice of USP1 targeting in the treatment of breast cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
03 February 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41388-022-02215-y
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30.
Weigelt B, Peterse JL, van ‘t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602.
Siegel MB, He X, Hoadley KA, Hoyle A, Pearce JB, Garrett AL, et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest. 2018;128:1371–83.
Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast. Cancer Cell. 2015;163:506–19.
Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5:1164–77.
Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005.
Brown D, Smeets D, Szekely B, Larsimont D, Szasz AM, Adnet PY, et al. Phylogenetic analysis of metastatic progression in breast cancer using somatic mutations and copy number aberrations. Nat Commun. 2017;8:14944.
Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–56.
Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57–78.
Xiao Z, Zhang P, Ma L. The role of deubiquitinases in breast cancer. Cancer Metastas Rev. 2016;35:589–600.
Wu Y, Wang Y, Lin Y, Liu Y, Wang Y, Jia J, et al. Dub3 inhibition suppresses breast cancer invasion and metastasis by promoting Snail1 degradation. Nat Commun. 2017;8:14228.
Zhang L, Zhou F, Drabsch Y, Gao R, Snaar-Jagalska BE, Mickanin C, et al. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-beta type I receptor. Nat Cell Biol. 2012;14:717–26.
Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, et al. Deubiquitylation and stabilization of PTEN by USP13. Nat Cell Biol. 2013;15:1486–94.
Fujiwara T, Saito A, Suzuki M, Shinomiya H, Suzuki T, Takahashi E, et al. Identification and chromosomal assignment of USP1, a novel gene encoding a human ubiquitin-specific protease. Genomics. 1998;54:155–8.
Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D’Andrea AD, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–9.
Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, Simpson LJ, et al. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell. 2007;28:798–809.
Ogrunc M, Martinez-Zamudio RI, Sadoun PB, Dore G, Schwerer H, Pasero P, et al. USP1 regulates cellular senescence by controlling genomic integrity. Cell Rep. 2016;15:1401–11.
Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–47.
Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, et al. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell. 2011;146:918–30.
Mistry H, Hsieh G, Buhrlage SJ, Huang M, Park E, Cuny GD, et al. Small-molecule inhibitors of USP1 target ID1 degradation in leukemic cells. Mol Cancer Ther. 2013;12:2651–62.
Das DS, Das A, Ray A, Song Y, Samur MK, Munshi NC, et al. Blockade of deubiquitylating enzyme USP1 inhibits DNA repair and triggers apoptosis in multiple myeloma cells. Clin Cancer Res. 2017;23:4280–9.
Lee JK, Chang N, Yoon Y, Yang H, Cho H, Kim E, et al. USP1 targeting impedes GBM growth by inhibiting stem cell maintenance and radioresistance. Neuro Oncol. 2016;18:37–47.
Liang Q, Dexheimer TS, Zhang P, Rosenthal AS, Villamil MA, You C, et al. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol. 2014;10:298–304.
Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–13.
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9.
Pau NiIB, Zakaria Z, Muhammad R, Abdullah N, Ibrahim N, Aina Emran N, et al. Gene expression patterns distinguish breast carcinomas from normal breast tissues: the Malaysian context. Pathol Res Pract. 2010;206:223–8.
Gluz O, Wild P, Meiler R, Diallo-Danebrock R, Ting E, Mohrmann S, et al. Nuclear karyopherin alpha2 expression predicts poor survival in patients with advanced breast cancer irrespective of treatment intensity. Int J Cancer. 2008;123:1433–8.
Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–82.
Altan B, Yokobori T, Mochiki E, Ohno T, Ogata K, Ogawa A, et al. Nuclear karyopherin-alpha2 expression in primary lesions and metastatic lymph nodes was associated with poor prognosis and progression in gastric cancer. Carcinogenesis. 2013;34:2314–21.
Jensen JB, Munksgaard PP, Sorensen CM, Fristrup N, Birkenkamp-Demtroder K, Ulhoi BP, et al. High expression of karyopherin-alpha2 defines poor prognosis in non-muscle-invasive bladder cancer and in patients with invasive bladder cancer undergoing radical cystectomy. Eur Urol. 2011;59:841–8.
Noetzel E, Rose M, Bornemann J, Gajewski M, Knuchel R, Dahl E. Nuclear transport receptor karyopherin-alpha2 promotes malignant breast cancer phenotypes in vitro. Oncogene. 2012;31:2101–14.
Mothi M, Sampson S. Pimozide for schizophrenia or related psychoses. Cochrane Database Syst Rev. 2013:CD001949.
Tueth MJ, Cheong JA. Clinical uses of pimozide. South Med J. 1993;86:344–9.
Chen J, Dexheimer TS, Ai Y, Liang Q, Villamil MA, Inglese J, et al. Selective and cell-active inhibitors of the USP1/ UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem Biol. 2011;18:1390–1400.
Yu Z, Song H, Jia M, Zhang J, Wang W, Li Q, et al. USP1-UAF1 deubiquitinase complex stabilizes TBK1 and enhances antiviral responses. J Exp Med. 2017;214:3553–63.
Raimondi M, Marcassa E, Cataldo F, Arnandis T, Mendoza-Maldonado R, Bestagno M, et al. Calpain restrains the stem cells compartment in breast cancer. Cell Cycle. 2016;15:106–16.
Sampieri K, Fodde R. Cancer stem cells and metastasis. Semin Cancer Biol. 2012;22:187–93.
Rudland PS, Platt-Higgins A, Renshaw C, West CR, Winstanley JH, Robertson L, et al. Prognostic significance of the metastasis-inducing protein S100A4 (p9Ka) in human breast cancer. Cancer Res. 2000;60:1595–603.
Moreno-Bueno G, Salvador F, Martin A, Floristan A, Cuevas EP, Santos V, et al. Lysyl oxidase-like 2 (LOXL2), a new regulator of cell polarity required for metastatic dissemination of basal-like breast carcinomas. EMBO Mol Med. 2011;3:528–44.
Ree AH, Florenes VA, Berg JP, Maelandsmo GM, Nesland JM, Fodstad O. High levels of messenger RNAs for tissue inhibitors of metalloproteinases (TIMP-1 and TIMP-2) in primary breast carcinomas are associated with development of distant metastases. Clin Cancer Res. 1997;3:1623–8.
Christiansen A, Dyrskjot L. The functional role of the novel biomarker karyopherin alpha 2 (KPNA2) in cancer. Cancer Lett. 2013;331:18–23.
Dankof A, Fritzsche FR, Dahl E, Pahl S, Wild P, Dietel M, et al. KPNA2 protein expression in invasive breast carcinoma and matched peritumoral ductal carcinoma in situ. Virchows Arch. 2007;451:877–81.
Kane RC, Farrell AT, Sridhara R, Pazdur R. United States Food and Drug Administration approval summary: bortezomib for the treatment of progressive multiple myeloma after one prior therapy. Clin Cancer Res. 2006;12:2955–60.
Nicholson B, Marblestone JG, Butt TR, Mattern MR. Deubiquitinating enzymes as novel anticancer targets. Future Oncol. 2007;3:191–9.
Rahme GJ, Zhang Z, Young AL, Cheng C, Bivona EJ, Fiering SN, et al. PDGF engages an E2F-USP1 signaling pathway to support ID2-mediated survival of proneural glioma cells. Cancer Res. 2016;76:2964–76.
Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11.
Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81572293, 31770976, and 81672359), Natural Science Foundation of Shanghai (18ZR1436800), the State Key Laboratory of Oncogenes and Related Genes (91–1705, 91-17-11), and Shanghai Cancer Institute (SB18-07).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Ma, A., Tang, M., Zhang, L. et al. USP1 inhibition destabilizes KPNA2 and suppresses breast cancer metastasis. Oncogene 38, 2405–2419 (2019). https://doi.org/10.1038/s41388-018-0590-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0590-8
This article is cited by
-
Emerging roles of deubiquitinating enzymes in actin cytoskeleton and tumor metastasis
Cellular Oncology (2024)
-
Silencing KPNA2 Promotes Ferroptosis in Laryngeal Cancer by Activating the FoxO Signaling Pathway
Biochemical Genetics (2024)
-
USP1 promotes cholangiocarcinoma progression by deubiquitinating PARP1 to prevent its proteasomal degradation
Cell Death & Disease (2023)
-
Ubiquitin-specific peptidase 1: assessing its role in cancer therapy
Clinical and Experimental Medicine (2023)
-
USP2 promotes cell proliferation and metastasis in choroidal melanoma via stabilizing Snail
Journal of Cancer Research and Clinical Oncology (2023)