Abstract
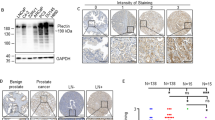
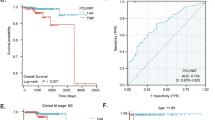
Most prostate cancer cases remain indolent for long periods of time, but metastatic progression quickly worsens the prognosis and leads to mortality. However, little is known about what promotes the metastasis of prostate cancer and there is a lack of effective prognostic indicators, making it immensely difficult to manage options for treatment or surveillance. Arginyltransferase 1 (Ate1) is the enzyme mediating post-translational protein arginylation, which has recently been identified as a master regulator affecting many cancer-relevant pathways including stress response, cell cycle checkpoints, and cell migration/adhesion. However, the precise role of Ate1 in cancer remains unknown. In this study, we found the occurrence of metastasis of prostate cancer is inversely correlated with the levels of Ate1 protein and mRNA in the primary tumor. We also found that metastatic prostate cancer cell lines have a reduced level of Ate1 protein compared to non-metastatic cell lines, and that a depletion of Ate1 drives prostate cancer cells towards more aggressive pro-metastatic phenotypes without affecting proliferation rates. Furthermore, we demonstrated that a reduction of Ate1 can result from chronic stress, and that shRNA-reduced Ate1 increases cellular resistance to stress, and drives spontaneous and stress-induced genomic mutations. Finally, by using a prostate orthotropic xenograft mouse model, we found that a reduction of Ate1 was sufficient to enhance the metastatic phenotypes of prostate cancer cell line PC-3 in vivo. Our study revealed a novel role of Ate1 in suppressing prostate cancer metastasis, which has a profound significance for establishing metastatic indicators for prostate cancer, and for finding potential treatments to prevent its metastasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol, Biomark Prev a Publ Am Assoc Cancer Res, cosponsored Am Soc Prev. 2016;25:16–27.
Jiang WG, Sanders AJ, Katoh M, Ungefroren H, Gieseler F, Prince M, et al. Tissue invasion and metastasis: molecular, biological and clinical perspectives. Semin Cancer Biol. 2015;35:S244–S75.
Klein CA. Selection and adaptation during metastatic cancer progression. Nature. 2013;501:365–72.
Kumar A, Birnbaum MD, Patel DM, Morgan WM, Singh J, Barrientos A. et al. Posttranslational arginylation enzyme Ate1 affects DNA mutagenesis by regulating stress response. Cell Death Dis. 2016;7:e2378
Eisenach PA, Schikora F, Posern G. Inhibition of arginyltransferase 1 induces transcriptional activity of myocardin-related transcription factor A (MRTF-A) and promotes directional migration. J Biol Chem. 2014;289:35376–87.
Zhang F, Saha S, Kashina A. Arginylation-dependent regulation of a proteolytic product of talin is essential for cell–cell adhesion. J Cell Biol. 2012;197:819–36.
Zhang N, Donnelly R, Ingoglia NA. Evidence that oxidized proteins are substrates for N-terminal arginylation. Neurochem Res. 1998;23:1411–20.
Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci Publ Protein. 2011;20:1298–345.
Klemperer NS, Pickart CM. Arsenite inhibits two steps in the ubiquitin-dependent proteolytic pathway. J Biol Chem. 1989;264:19245–52.
Wong CC, Xu T, Rai R, Bailey AO, Yates JR,3rd. et al. Global analysis of posttranslational protein arginylation. PLoS Biol. 2007;5:e258
Piatkov KI, Brower CS, Varshavsky A. The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc Natl Acad Sci USA. 2012;109:E1839–47.
Decca MB, Bosc C, Luche S, Brugiere S, Job D, Rabilloud T, et al. Protein arginylation in rat brain cytosol: a proteomic analysis. Neurochem Res. 2006;31:401–9.
Balzi E, Choder M, Chen WN, Varshavsky A, Goffeau A. Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J Biol Chem. 1990;265:7464–71.
Kwon YT, Kashina AS, Varshavsky A. Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol Cell Biol. 1999;19:182–93.
Hu RG, Brower CS, Wang H, IV Davydov, J Sheng, J Zhou, et al. Arginyltransferase, its specificity, putative substrates, bidirectional promoter, and splicing-derived isoforms. J Biol Chem. 2006;281:32559–73.
Rai R, Mushegian A, Makarova K, Kashina A. Molecular dissection of arginyltransferases guided by similarity to bacterial peptidoglycan synthases. EMBO Rep. 2006;7:800–5.
Rai R, Zhang F, Colavita K, Leu NA, Kurosaka S, Kumar A, et al. Arginyltransferase suppresses cell tumorigenic potential and inversely correlates with metastases in human cancers. Oncogene. 2016;35:4058–68.
Kumar B, Koul S, Khandrika L, Meacham RB, Koul HK. Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer Res. 2008;68:1777–85.
Weinstein JN, Collisson EA, Mills GB, Shaw KM, Ozenberger BA, Ellrott K, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–20.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22.
Rusthoven CG, Carlson JA, Waxweiler TV, Yeh N, Raben D, Flaig TW, et al. The prognostic significance of Gleason scores in metastatic prostate cancer. Urol Oncol. 2014;32:707–13.
Kuroiwa K, Uchino H, Yokomizo A, Naito S. Impact of reporting rules of biopsy Gleason score for prostate cancer. J Clin Pathol. 2009;62:260–3.
Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45.
Suzuki F, Suzuki K, Nikaido O. An improved soft agar method for determining neoplastic transformation in vitro. J Tissue Cult Methods. 1983;8:109–13.
Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–33.
Albini A, Benelli R. The chemoinvasion assay: a method to assess tumor and endothelial cell invasion and its modulation. Nat Protoc. 2007;2:504–11.
Rice MA, Ishteiwy RA, Magani F, Udayakumar T, Reiner T, Yates TJ, et al. The microRNA-23b/-27b cluster suppresses prostate cancer metastasis via Huntingtin-interacting protein 1-related. Oncogene. 2016;35:4752–61.
Wang M, Stearns ME. Isolation and characterization of PC-3 human prostatic tumor sublines which preferentially metastasize to select organs in S.C.I.D. mice. Differ; Res Biol Divers. 1991;48:115–25.
Zhang F, Saha S, Shabalina SA, Kashina A. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science. 2010;329:1534–7.
Rai R, Zhang F, Colavita K, Leu NA, Kurosaka S, Kumar A. et al. Arginyltransferase suppresses cell tumorigenic potential and inversely correlates with metastases in human cancers. Oncogene. 2016;35:4058–68.
Fidler IJ. Tumor heterogeneity and the biology of cancer invasion and metastasis. Cancer Res. 1978;38:2651–60.
de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-beta signaling in cancer. J Natl Cancer Inst. 2000;92:1388–402.
Sherman MY, Gabai VL. Hsp70 in cancer: back to the future. Oncogene. 2015;34:4153–61.
Massague J. TGFbeta in cancer. Cell. 2008;134:215–30.
Humphrey PA. Gleason grading and prognostic factors in carcinoma of the prostate. Mod Pathol. 2004;17:292–306.
Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323.
Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinforma. 2010;11:94.
Kurosaka S, Leu NA, Zhang F, Bunte R, Saha S, Wang J. et al. Arginylation-dependent neural crest cell migration is essential for mouse development. PLoS Genet. 2010;6:e1000878
Acknowledgements
Cell sorting was performed in the core facility of Sylvester Comprehensive Cancer Center. Ex vivo imaging was performed in the small animal imaging facility at the University of Miami Miller School of medicine. Gamma irradiation was performed in the Radiation Control core facility at the University of Miami Miller School of medicine. Immunostaining of tumor array samples was performed in the histology core facility in the Hussman Institute for Human Genomics at the University of Miami Miller School of medicine. Dr. Priyamvada Rai (University of Miami) provided critical reading for the manuscript. Dr. Yuanfang Guan (University of Michigan) provided technical consultations for the analysis related to Ate1 mRNA levels. The research presented in this article is funded in part by these grants: DoD (CDMRP), Idea Award, PC140622; DoD (CDMRP), Exploratory grant, PC141013; and NIGMS (NIH), R01GM107333.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Birnbaum, M.D., Zhao, N., Moorthy, B.T. et al. Reduced Arginyltransferase 1 is a driver and a potential prognostic indicator of prostate cancer metastasis. Oncogene 38, 838–851 (2019). https://doi.org/10.1038/s41388-018-0462-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0462-2
This article is cited by
-
MiR-183-5p promotes migration and invasion of prostate cancer by targeting TET1
BMC Urology (2023)
-
Inhibition of cancer cell-derived exosomal microRNA-183 suppresses cell growth and metastasis in prostate cancer by upregulating TPM1
Cancer Cell International (2021)