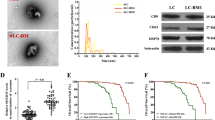
Abstract
Multiple myeloma (MM) is characterized by the decreased osteogenic potential of mesenchymal stem cells (MSCs). Communication between cancer cells and cancer stromal cells is a driving factor in tumor progression. Understanding the myeloma–stroma interactions is critical to the development of effective strategies that can reverse bone diseases. Here we identified that bioactive lncRNA RUNX2-AS1 in myeloma cells could be packed into exosomes and transmitted to MSCs, thus repressing the osteogenesis of MSCs. RUNX2-AS1, which arises from the antisense strand of RUNX2, was enriched in MSCs derived from MM patients (MM-MSCs). RUNX2-AS1 was capable of forming an RNA duplex with RUNX2 pre-mRNA at overlapping regions and this duplex transcriptionally repressed RUNX2 expression by reducing the splicing efficiency, resulting in decreased osteogenic potential of MSCs. In vivo mouse models, administered an inhibitor of exosome secretion, GW4869, was found to be effective in preventing bone loss, sustained by both bone formation and anticatabolic activities. Therefore, exosomal lncRNA RUNX2-AS1 may serve as a potential therapeutic target for bone lesions in MM. In summary, our results indicated a key role of exosomal lncRUNX2-AS1 in transferring from MM cells to MSCs in osteogenic differentiation, through a unique exosomal lncRUNX2-AS1/RUNX2 pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Giuliani N, Rizzoli V, Roodman GD. Multiple myeloma bone disease: pathophysiology of osteoblast inhibition. Blood. 2006;108:3992–6.
Raje N, Roodman GD. Advances in the biology and treatment of bone disease in multiple myeloma. Clin Cancer Res. 2011;17:1278–86.
Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–68.
Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159:499–513.
Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T, et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst. 2015;107:djv135.
Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–15.
Roccaro AM, Sacco A, Maiso P, Azab AK, Tai YT, Reagan M, et al. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J Clin Invest. 2013;123:1542–55.
Xin H, Li Y, Buller B, Katakowski M, Zhang Y, Wang X, et al. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells. 2012;30:1556–64.
Lokody I. Cancer genetics: exosomally derived miR-105 destroys tight junctions. Nat Rev Genet. 2014;15:362.
Luga V, Wrana JL. Tumor-stroma interaction: Revealing fibroblast-secreted exosomes as potent regulators of Wnt-planar cell polarity signaling in cancer metastasis. Cancer Res. 2013;73:6843–7.
Gezer U, Ozgur E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38:1076–9.
Mohankumar S, Patel T. Extracellular vesicle long noncoding RNA as potential biomarkers of liver cancer. Brief Funct Genom. 2015;15:249–56.
Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007–12.
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–25.
He C, Sidoli S, Warneford-Thomson R, Tatomer DC, Wilusz JE, Garcia BA, et al. High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol Cell. 2016;64:416–30.
Hudson WH, Pickard MR, de Vera IM, Kuiper EG, Mourtada-Maarabouni M, Conn GL, et al. Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate. Nat Commun. 2014;5:5395.
Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46.
Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–43.
Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9.
Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–7.
Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013;14:319.
Takahashi K, Yan IK, Wood J, Haga H, Patel T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol Cancer Res. 2014;12:1377–87.
Franceschi RT, Xiao G, Jiang D, Gopalakrishnan R, Yang S, Reith E. Multiple signaling pathways converge on the Cbfa1/Runx2 transcription factor to regulate osteoblast differentiation. Connect Tissue Res. 2003;44(Suppl 1):109–16.
Komori T. Regulation of skeletal development by the Runx family of transcription factors. J Cell Biochem. 2005;95:445–53.
Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6.
Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–8.
He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–7.
Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Akiyama M, et al. Inhibition of the insulin-like growth factor receptor-1 tyrosine kinase activity as a therapeutic strategy for multiple myeloma, other hematologic malignancies, and solid tumors. Cancer Cell. 2004;5:221–30.
Fabbri M,Paone A,Calore F,Galli R,Gaudio E,Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110–6.
Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–52.
Zhuang W, Ge X, Yang S, Huang M, Zhuang W, Chen P, et al. Upregulation of lncRNA MEG3 promotes osteogenic differentiation of mesenchymal stem cells from multiple myeloma patients by targeting BMP4 transcription. Stem Cells. 2015;33:1985–97.
Li B, Shi M, Li J, Zhang H, Chen B, Chen L, et al. Elevated tumor necrosis factor-alpha suppresses TAZ expression and impairs osteogenic potential of Flk-1+mesenchymal stem cells in patients with multiple myeloma. Stem Cells Dev. 2007;16:921–30.
Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC, Tsai PH, et al. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36:4929–42.
Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun. 2015;6:6999.
Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–81.
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–3.
Scarola M, Comisso E, Pascolo R, Chiaradia R, Marion RM, Schneider C, et al. Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat Commun. 2015;6:7631.
Lister N, Shevchenko G, Walshe JL, Groen J, Johnsson P, Vidarsdottir L, et al. The molecular dynamics of long noncoding RNA control of transcription in PTEN and its pseudogene. Proc Natl Acad Sci USA. 2017;114:9942–7.
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;2:666–81.
Saldana-Meyer R, Gonzalez-Buendia E, Guerrero G, Narendra V, Bonasio R, Recillas-Targa F, et al. CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes & Dev. 2014;28:723–34.
Guo J, Cheng P, Yuan H, Liu Y. The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell. 2009;138:1236–46.
Shah S, Wittmann S, Kilchert C, Vasiljeva L. lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev. 2014;28:231–44.
Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MA, Sadek P, Sie D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–58.
Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife. 2016;5:e19276.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Acknowledgements
This work was supported in part by National Natural Science Foundation of China (81673448, 81670191), Natural Science Foundation of Jiangsu Province China (BK 20161218, BK20161223), The Applied Basic Research Programs of Suzhou City (SYS201546), and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
These authors contributed equally: Bingzong Li and Hongxia Xu.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Li, B., Xu, H., Han, H. et al. Exosome-mediated transfer of lncRUNX2-AS1 from multiple myeloma cells to MSCs contributes to osteogenesis. Oncogene 37, 5508–5519 (2018). https://doi.org/10.1038/s41388-018-0359-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0359-0
This article is cited by
-
The role and applications of extracellular vesicles in osteoporosis
Bone Research (2024)
-
Unveiling the functional paradigm of exosome-derived long non-coding RNAs (lncRNAs) in cancer: based on a narrative review and systematic review
Journal of Cancer Research and Clinical Oncology (2023)
-
Identification of the key exosomal lncRNAs/mRNAs in the serum during distraction osteogenesis
Journal of Orthopaedic Surgery and Research (2022)
-
Molecular functions and therapeutic applications of exosomal noncoding RNAs in cancer
Experimental & Molecular Medicine (2022)
-
An exosome-related long non-coding RNAs risk model could predict survival outcomes in patients with breast cancer
Scientific Reports (2022)