Abstract
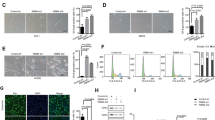
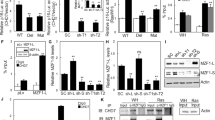
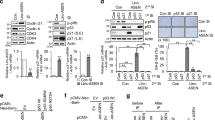
miR-30 is a microRNA frequently overexpressed in human cancers. However, the biological consequence of miR-30 overexpression in cancer has been unclear. In a genetic screen, miR-30 was found to abrogate oncogenic-induced senescence, a key tumor-suppressing mechanism that involves DNA damage responses, activation of p53 and induction of p16INK4A. In cells and mouse models, miR-30 disrupts senescence and promotes cancer by suppressing 2 targets, CHD7 and TNRC6A. We show that while CHD7 is a transcriptional coactivator essential for induction of p16INK4A in senescent cells, TNRC6A, a miRNA machinery component, is required for expression and functionality of DNA damage response RNAs (DDRNAs) that mediate DNA damage responses and p53 activation by orchestrating histone modifications, chromatin remodeling and recruitment of DNA damage factors at damaged sites. Thus, miR-30 inhibits both p16INK4A and p53, 2 key senescence effectors, leading to efficient senescence disruption. These findings have identified novel signaling pathways mediating oncogene-induced senescence and tumor-suppression, and revealed the molecular and cellular mechanisms underlying the oncogenic activity of miR-30. Thus, the miR-30/CHD7/TNRC6A pathway is potentially a novel diagnostic biomarker and therapeutic target for cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008;14:1–6.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, et al. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–96.
Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27:2801–9.
Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–7.
Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–42.
Wang W, Chen JX, Liao R, Deng Q, Zhou JJ, Huang S, et al. Sequential activation of the MEK-extracellular signal-regulated kinase and MKK3/6-p38 mitogen-activated protein kinase pathways mediates oncogenic ras-induced premature senescence. Mol Cell Biol. 2002;22:3389–403.
Kwong J, Hong L, Liao R, Deng Q, Han J, Sun P. p38alpha and p38gamma mediate oncogenic ras-induced senescence through differential mechanisms. J Biol Chem. 2009;284:11237–46.
Kwong J, Chen M, Lv D, Luo N, Su W, Xiang R, et al. Induction of p38delta expression plays an essential role in oncogenic ras-induced senescence. Mol Cell Biol. 2013;33:3780–94.
Ferbeyre G, de Stanchina E, Lin AW, Querido E, McCurrach ME, Hannon GJ, et al. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol Cell Biol. 2002;22:3497–508.
Narita M, Lowe SW. Senescence comes of age. Nat Med. 2005;11:920–2.
Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, et al. PRAK is essential for ras-induced senescence and tumor suppression. Cell. 2007;128:295–308.
Zhao T, Li J, Chen AF. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am J Physiol Endocrinol Metab. 2010;299:E110–6.
Faraonio R, Salerno P, Passaro F, Sedia C, Iaccio A, Bellelli R, et al. A set of miRNAs participates in the cellular senescence program in human diploid fibroblasts. Cell Death Differ. 2011;19:713–21.
Srikantan S, Gorospe M, Abdelmohsen K. Senescence-associated microRNAs linked to tumorigenesis. Cell Cycle. 2011;10:3211–2.
Hong L, Lai M, Chen M, Xie C, Liao R, Kang YJ, et al. The miR-17-92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010;70:8547–57.
Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, et al. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–12.
Francia S, Michelini F, Saxena A, Tang D, de HM, Anelli V, et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–5.
Wang Q, Goldstein M. Small RNAs recruit chromatin-modifying enzymes MMSET and Tip60 to reconfigure damaged DNA upon double-strand break and facilitate repair. Cancer Res. 2016;76:1904–15.
Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–59.
DiGiovanni J. Modification of multistage skin carcinogenesis in mice. Prog Exp Tumor Res. 1991;33:192–229.
DiGiovanni J. Multistage carcinogenesis in mouse skin. Pharmacol Ther. 1992;54:63–128.
Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–50.
Caldwell ME, DeNicola GM, Martins CP, Jacobetz MA, Maitra A, Hruban RH, et al. Cellular features of senescence during the evolution of human and murine ductal pancreatic cancer. Oncogene. 2012;31:1599–608.
Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–5.
Schnetz MP, Bartels CF, Shastri K, Balasubramanian D, Zentner GE, Balaji R, et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601.
Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6:e1001023.
Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–62.
Engelen E, Akinci U, Bryne JC, Hou J, Gontan C, Moen M, et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43:607–11.
Kim MS, Oh JE, Kim YR, Park SW, Kang MR, Kim SS, et al. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. J Pathol. 2010;221:139–46.
Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965.
Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–4.
Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7.
d’Adda di FF. A direct role for small non-coding RNAs in DNA damage response. Trends Cell Biol. 2014;24:171–8.
Wang H, Shao Z, Shi LZ, Hwang PY, Truong LN, Berns MW, et al. CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J Biol Chem. 2012;287:21471–80.
Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell. 2012;48:723–33.
Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152:1344–54.
Berkovich E, Monnat RJ Jr., Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–90.
Hurd EA, Capers PL, Blauwkamp MN, Adams ME, Raphael Y, Poucher HK, et al. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm Genome. 2007;18:94–104.
Jongmans MC, Admiraal RJ, van der Donk KP, Vissers LE, Baas AF, Kapusta L, et al. CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. J Med Genet. 2006;43:306–14.
Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–51.
Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, et al. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–55.
Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–59.
Zamudio JR, Kelly TJ, Sharp PA. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell. 2014;156:920–34.
Sun P, Dong P, Dai K, Hannon GJ, Beach D. p53-independent role of MDM2 in TGF-beta1 resistance. Science. 1998;282:2270–2.
Zeng PY,Vakoc CR,Chen ZC,Blobel GA,Berger SL, In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques. 2006;41:694–6.
Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc. 2008;3:698–709.
Gertsenstein M, Nutter LM, Reid T, Pereira M, Stanford WL, Rossant J, et al. Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PLoS ONE. 2010;5:e11260.
Yoshizuka N, Chen RM, Xu Z, Liao R, Hong L, Hu WY, et al. A novel function of p38-regulated/activated kinase in endothelial cell migration and tumor angiogenesis. Mol Cell Biol. 2012;32:606–18.
Acknowledgements
We thank Dr. Joanna Wysocka for CHD7 cDNA, and Tumor Tissue and Pathology, Biostatistics and Bioinformatics, Cell & Viral Vector Laboratory and Cellular Imaging Shared Resources of WFBCCC. This study was supported by Overseas Collaboration Fund from National NSF of China 31428013 (PS, RX), NIH/NCI grants CA106768, CA131231, CA172115 (PS), P30CA012197, and Thomas K. Hearn Brain Tumor Center. WS is supported by International Postdoctoral Exchange Fellowship Program by Office of China Postdoctoral Council (20140027).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
These authors contribute equally: Weijun Su, Lixin Hong, Xin Xu.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Su, W., Hong, L., Xu, X. et al. miR-30 disrupts senescence and promotes cancer by targeting both p16INK4A and DNA damage pathways. Oncogene 37, 5618–5632 (2018). https://doi.org/10.1038/s41388-018-0358-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0358-1
This article is cited by
-
LINC01082 Inhibits Non-Small Cell Lung Cancer by Targeting the miR-543/TNRC6A Axis
Biochemical Genetics (2023)
-
MZF1 mediates oncogene-induced senescence by promoting the transcription of p16INK4A
Oncogene (2022)
-
Revisiting cancer hallmarks: insights from the interplay between oxidative stress and non-coding RNAs
Molecular Biomedicine (2020)
-
The roles of MTOR and miRNAs in endothelial cell senescence
Biogerontology (2020)
-
miRNAs expression signature potentially associated with lymphatic dissemination in locally advanced prostate cancer
BMC Medical Genomics (2020)