Abstract
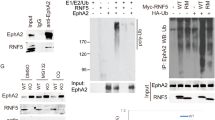
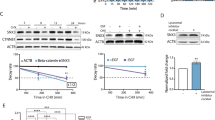
The RANK/RANKL axis emerges as a key regulator of breast cancer initiation, progression, and metastasis. RANK-c is a RANK receptor isoform produced through alternative splicing of the TNFRSF11A (RANK) gene and a dominant-negative regulator of RANK-induced nuclear factor-κB (NF-κB) activation. Here we report that RANK-c transcript is expressed in 3.2% of cases in The Cancer Genome Atlas breast cancer cohort evenly between ER-positive and ER-negative cases. Nevertheless, the ratio of RANK to RANK-c (RANK/RANK-c) is increased in ER-negative breast cancer cell lines compared to ER-positive breast cancer cell lines. In addition, forced expression of RANK-c in ER-negative breast cancer cell lines inhibited stimuli-induced NF-κB activation and attenuated migration, invasion, colony formation, and adhesion of cancer cells. Further, RANK-c expression in MDA-MB-231 cells inhibited lung metastasis and colonization in vivo. The RANK-c-mediated inhibition of cancer cell aggressiveness and nuclear factor-κB (NF-κB) activation in breast cancer cells seems to rely on a RANK-c/TNF receptor-associated factor-2 (TRAF2) protein interaction. This was further confirmed by a mutated RANK-c that is unable to interact with TRAF2 and abolishes the ability to attenuate NF-κB activation, migration, and invasion. Additional protein interaction characterization revealed epidermal growth factor receptor (EGFR) as a novel interacting partner for RANK-c in breast cancer cells with a negative effect on EGFR phosphorylation and EGF-dependent downstream signaling pathway activation. Our findings further elucidate the complex molecular biology of the RANKL/RANK system in breast cancer and provide preliminary data for RANK-c as a possible marker for disease progression and aggressiveness.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Papanastasiou AD, Sirinian C, Kalofonos HP, Repanti M. Current therapeutic leads for the treatment of melanoma: targeted immunotherapy in the post-genomic era. Curr Mol Pharmacol. 2014;7:33–43.
Lopez-Garcia MA, Geyer FC, Lacroix-Triki M, Marchio C, Reis-Filho JS. Breast cancer precursors revisited: molecular features and progression pathways. Histopathology. 2010;57:171–92.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800.
Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010;220:263–80.
Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6:718–30.
Lacroix M, Toillon RA, Leclercq G. Stable ‘portrait’ of breast tumors during progression: data from biology, pathology and genetics. Endocr-Relat Cancer. 2004;11:497–522.
Loo LW, Grove DI, Williams EM, Neal CL, Cousens LA, Schubert EL, et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64:8541–9.
Melchor L, Honrado E, Garcia MJ, Alvarez S, Palacios J, Osorio A, et al. Distinct genomic aberration patterns are found in familial breast cancer associated with different immunohistochemical subtypes. Oncogene. 2008;27:3165–75.
Rennstam K, Ahlstedt-Soini M, Baldetorp B, Bendahl PO, Borg A, Karhu R, et al. Patterns of chromosomal imbalances defines subgroups of breast cancer with distinct clinical features and prognosis. A study of 305 tumors by comparative genomic hybridization. Cancer Res. 2003;63:8861–8.
Schramek D, Leibbrandt A, Sigl V, Kenner L, Pospisilik JA, Lee HJ, et al. Osteoclast differentiation factor RANKL controls development of progestin-driven mammary cancer. Nature. 2010;468:98–102.
Gonzalez-Suarez E, Jacob AP, Jones J, Miller R, Roudier-Meyer MP, Erwert R, et al. RANK ligand mediates progestin-induced mammary epithelial proliferation and carcinogenesis. Nature. 2010;468:103–7.
Jones DH, Nakashima T, Sanchez OH, Kozieradzki I, Komarova SV, Sarosi I, et al. Regulation of cancer cell migration and bone metastasis by RANKL. Nature. 2006;440:692–6.
Pfitzner BM, Branstetter D, Loibl S, Denkert C, Lederer B, Schmitt WD, et al. RANK expression as a prognostic and predictive marker in breast cancer. Breast Cancer Res Treat. 2014;145:307–15.
Tan W, Zhang W, Strasner A, Grivennikov S, Cheng JQ, Hoffman RM, et al. Tumour-infiltrating regulatory T cells stimulate mammary cancer metastasis through RANKL-RANK signalling. Nature. 2011;470:548–53.
Papanastasiou AD, Sirinian C, Kalofonos HP. Identification of novel human receptor activator of nuclear factor-kB isoforms generated through alternative splicing: implications in breast cancer cell survival and migration. Breast Cancer Res. 2012;14:R112.
Sirinian C, Papanastasiou AD, Zarkadis IK, Kalofonos HP. Alternative splicing generates a truncated isoform of human TNFRSF11A (RANK) with an altered capacity to activate NF-kappaB. Gene. 2013;525:124–9.
Galibert L, Tometsko ME, Anderson DM, Cosman D, Dougall WC. The involvement of multiple tumor necrosis factor receptor (TNFR)-associated factors in the signaling mechanisms of receptor activator of NF-kappaB, a member of the TNFR superfamily. J Biol Chem. 1998;273:34120–7.
Palafox M, Ferrer I, Pellegrini P, Vila S, Hernandez-Ortega S, Urruticoechea A, et al. RANK induces epithelial–mesenchymal transition and stemness in human mammary epithelial cells and promotes tumorigenesis and metastasis. Cancer Res. 2012;72:2879–88.
Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–37.
Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16.
Matsuda N, Lim B, Wang X, Ueno NT. Early clinical development of epidermal growth factor receptor targeted therapy in breast cancer. Expert Opin Investig Drugs. 2017;26:463–79.
Mendelsohn J, Baselga J. Epidermal growth factor receptor targeting in cancer. Semin Oncol. 2006;33:369–85.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Krikos A, Laherty CD, Dixit VM. Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J Biol Chem. 1992;267:17971–6.
Tanaka Y, Hayashi M, Takagi S, Yoshie O. Differential transactivation of the intercellular adhesion molecule 1 gene promoter by Tax1 and Tax2 of human T-cell leukemia viruses. J Virol. 1996;70:8508–17.
Ohtsuka T, Kubota A, Hirano T, Watanabe K, Yoshida H, Tsurufuji M, et al. Glucocorticoid-mediated gene suppression of rat cytokine-induced neutrophil chemoattractant CINC/gro, a member of the interleukin-8 family, through impairment of NF-kappa B activation. J Biol Chem. 1996;271:1651–9.
Kim HH, Lee DE, Shin JN, Lee YS, Jeon YM, Chung CH, et al. Receptor activator of NF-kappaB recruits multiple TRAF family adaptors and activates c-Jun N-terminal kinase. FEBS Lett. 1999;443:297–302.
Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–5.
Takeuchi M, Rothe M, Goeddel DV. Anatomy of TRAF2. Distinct domains for nuclear factor-kappaB activation and association with tumor necrosis factor signaling proteins. J Biol Chem. 1996;271:19935–42.
Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308.
Blakely CM, Pazarentzos E, Olivas V, Asthana S, Yan JJ, Tan I, et al. NF-kappaB-activating complex engaged in response to EGFR oncogene inhibition drives tumor cell survival and residual disease in lung cancer. Cell Rep. 2015;11:98–110.
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123:725–31.
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6.
Forootan SS, Butler JM, Gardener D, Baird AE, Dodson A, Darby A, et al. Transcriptome sequencing of human breast cancer reveals aberrant intronic transcription in amplicons and dysregulation of alternative splicing with major therapeutic implications. Int J Oncol. 2016;48:130–44.
Eswaran J, Horvath A, Godbole S, Reddy SD, Mudvari P, Ohshiro K, et al. RNA sequencing of cancer reveals novel splicing alterations. Sci Rep. 2013;3:1689.
Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7:e1002218.
Sebestyen E, Zawisza M, Eyras E. Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res. 2015;43:1345–56.
Biswas DK, Shi Q, Baily S, Strickland I, Ghosh S, Pardee AB, et al. NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc Natl Acad Sci USA. 2004;101:10137–42.
Singh S, Shi Q, Bailey ST, Palczewski MJ, Pardee AB, Iglehart JD, et al. Nuclear factor-kappaB activation: a molecular therapeutic target for estrogen receptor-negative and epidermal growth factor receptor family receptor-positive human breast cancer. Mol Cancer Ther. 2007;6:1973–82.
Nolan E, Vaillant F, Branstetter D, Pal B, Giner G, Whitehead L, et al. RANK ligand as a potential target for breast cancer prevention in BRCA1-mutation carriers. Nat Med. 2016;22:933–9.
Sigl V, Owusu-Boaitey K, Joshi PA, Kavirayani A, Wirnsberger G, Novatchkova M, et al. RANKL/RANK control Brca1 mutation-driven mammary tumors. Cell Res. 2016;26:761–74.
Yoldi G, Pellegrini P, Trinidad EM, Cordero A, Gomez-Miragaya J, Serra-Musach J, et al. RANK signaling blockade reduces breast cancer recurrence by inducing tumor cell differentiation. Cancer Res. 2016;76:5857–69.
Marazioti A, Kairi CA, Spella M, Giannou AD, Magkouta S, Giopanou I, et al. Beneficial impact of CCL2 and CCL12 neutralization on experimental malignant pleural effusion. PLoS ONE. 2013;8:e71207.
Giannou AD, Marazioti A, Spella M, Kanellakis NI, Apostolopoulou H, Psallidas I, et al. Mast cells mediate malignant pleural effusion formation. J Clin Investig. 2015;125:2317–34.
Acknowledgements
We thank the Advanced Light Microscopy facility of the Medical School, University of Patras, Greece. We also thank Dr. Ross (Department of Biochemistry and Molecular Pharmacology, University of Massachusetts Medical School, MA 01605, USA) for plasmid MSCV-XZ066-EGFRvIII (Addgene #20737).
Funding
CS is funded by the State Scholarships Foundation “IKY Fellowships of Excellence for Postgraduate Studies in Greece-Siemens Programme” (SR 229101). The research reported was partially funded by the State Scholarships Foundation (SR 229101). The publicly available dataset from the TCGA breast cancer study was retrieved from the TCGA data portal on 05/2017.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Sirinian, C., Papanastasiou, A.D., Schizas, M. et al. RANK-c attenuates aggressive properties of ER-negative breast cancer by inhibiting NF-κB activation and EGFR signaling. Oncogene 37, 5101–5114 (2018). https://doi.org/10.1038/s41388-018-0324-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0324-y
This article is cited by
-
TRAF6 as a potential target in advanced breast cancer: a systematic review, meta-analysis, and bioinformatics validation
Scientific Reports (2023)
-
Estrogen Receptor and the Unfolded Protein Response: Double-Edged Swords in Therapy for Estrogen Receptor-Positive Breast Cancer
Targeted Oncology (2022)
-
MCPIP1-mediated NFIC alternative splicing inhibits proliferation of triple-negative breast cancer via cyclin D1-Rb-E2F1 axis
Cell Death & Disease (2021)
-
RANK signaling increases after anti-HER2 therapy contributing to the emergence of resistance in HER2-positive breast cancer
Breast Cancer Research (2021)
-
RANKL/RANK/OPG system beyond bone remodeling: involvement in breast cancer and clinical perspectives
Journal of Experimental & Clinical Cancer Research (2019)