Abstract
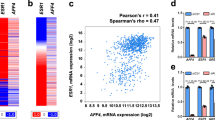
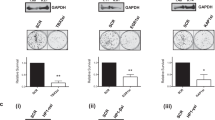
The chromatin state is finely tuned to regulate function and specificity for transcription factors such as oestrogen receptor alpha (ER), which contributes to cell growth in breast cancer. ER transcriptional potential is mediated, in large part, by the specific associated proteins and co-factors that interact with it. Despite the identification and characterisation of several ER coregulators, a complete and systematic view of ER-regulating chromatin modifiers is lacking. By exploiting a focused siRNA screen that investigated the requirement for a library of 330 chromatin regulators in ER-mediated cell growth, we find that the NuRD and coREST histone deacetylation complexes are critical for breast cancer cell proliferation. Further, by proteomic and genomics approaches, we discover the transcription factor TRPS1 to be a key interactor of the NuRD and coREST complexes. Interestingly, TRPS1 gene amplification occurs in 28% of human breast tumours and is associated with poor prognosis. We propose that TRPS1 is required to repress spurious binding of ER, where it contributes to the removal of histone acetylation. Our data suggest that TRPS1 is an important ER-associated transcriptional repressor that regulates cell proliferation, chromatin acetylation and ER binding at the chromatin of cis-regulatory elements.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Johnston SRD, Dowsett M. Aromatase inhibitors for breast cancer: lessons from the laboratory. Nat Rev Cancer. 2003;3:821–31.
Thomas C, Gustafsson J-Å. The different roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer. 2011;11:597–608.
Thomas C, Gustafsson J-Å. Progesterone receptor-estrogen receptor crosstalk: a novel insight. Trends Endocrinol Metab. 2015;26:453–4.
Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–9.
Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27.
Jozwik KM, Carroll JS. Pioneer factors in hormone-dependent cancers. Nat Rev Cancer. 2012;12:381–5.
Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, Ali S, Chin S-F, Palmieri C, Caldas C, Carroll JS. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–93.
Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet. 2013;14:765–80.
Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, METABRIC Group, Langerød A, Green A, Provenzano E, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52.
Jozwik KM, Chernukhin I, Serandour AA, Nagarajan S, Carroll JS. FOXA1 directs H3K4 monomethylation at enhancers via recruitment of the methyltransferase MLL3. Cell Rep. 2016;17:2715–23.
Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Métivier R, Salbert G, Eeckhoute J. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21:555–65.
Belandia B, Orford RL, Hurst HC, Parker MG. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 2002;21:4094–103.
DiRenzo J, Shang Y, Phelan M, Sif S, Myers M, Kingston R, Brown M. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol Cell Biol. 2000;20:7541–9.
Hohmann AF, Vakoc CR. A rationale to target the SWI/SNF complex for cancer therapy. Trends Genet. 2014;30:356–63.
Helin K, Dhanak D. Chromatin proteins and modifications as drug targets. Nature. 2013;502:480–8.
Fortunati N, Bertino S, Costantino L, De Bortoli M, Compagnone A, Bandino A, Catalano MG, Boccuzzi G. Valproic acid restores ER alpha and antiestrogen sensitivity to ER alpha-negative breast cancer cells. Mol Cell Endocrinol. 2010;314:17–22.
Alluri PG, Asangani IA, Chinnaiyan AM. BETs abet Tam-R in ER-positive breast cancer. Cell Res. 2014;24:899–900.
Ladd B, Mazzola AM, Bihani T, Lai Z, Bradford J, Collins M, Barry E, Goeppert AU, Weir HM, Hearne K, Renshaw JG, Mohseni M, Hurt E, Jalla S, Bao H, Hollingsworth R, Reimer C, Zinda M, Fawell S, D’Cruz CM. Effective combination therapies in preclinical endocrine resistant breast cancer models harboring ER mutations. Oncotarget. 2016;7:54120–36.
Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Huh SJ, Liang Y, Ryan J, Doherty E, Mohammed H, Guo H, Stover DG, Ekram MB, Peluffo G, Brown J, D’Santos C, Krop IE, Dillon D, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529:413–7.
Mohammed H, D’Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, Rueda OM, Holmes KA, Theodorou V, Robinson JLL, Zwart W, Saadi A, Ross-Innes CS, Chin S-F, Menon S, Stingl J, Palmieri C, Caldas C, Carroll JS. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–9.
Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J, Sirokmány G, Donati G, Uribe-Lewis S, Pavlidis P, Murrell A, Markowetz F, Watt FM. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol. 2012;14:753–63.
Kawai H, Li H, Chun P, Avraham S, Avraham HK. Direct interaction between BRCA1 and the estrogen receptor regulates vascular endothelial growth factor (VEGF) transcription and secretion in breast cancer cells. Oncogene. 2002;21:7730–9.
Nagarajan S, Hossan T, Alawi M, Najafova Z, Indenbirken D, Bedi U, Taipaleenmäki H, Ben-Batalla I, Scheller M, Loges S, Knapp S, Hesse E, Chiang C-M, Grundhoff A, Johnsen SA. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep. 2014;8:460–9.
Toska E, Osmanbeyoglu HU, Castel P, Chan C, Hendrickson RC, Elkabets M, Dickler MN, Scaltriti M, Leslie CS, Armstrong SA, Baselga J. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science. 2017;355:1324–30.
Mohammed H, Taylor C, Brown GD, Papachristou EK, Carroll JS, D’Santos CS. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11:316–26.
Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 2013;23:12–22.
Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–83.
Perdomo J, Crossley M. The Ikaros family protein Eos associates with C-terminal-binding protein corepressors. Eur J Biochem. 2002;269:5885–92.
Wu L, Wang Y, Liu Y, Yu S, Xie H, Shi X, Qin S, Ma F, Tan TZ, Thiery JP, Chen L. A central role for TRPS1 in the control of cell cycle and cancer development. Oncotarget. 2014;5:7677–90.
Chen JQ, Bao Y, Litton J, Xiao L, Zhang H-Z, Warneke CL, Wu Y, Shen X, Wu S, Katz RL, Sahin A, Bondy M, Murray JL, Radvanyi L. Expression and relevance of TRPS-1: a new GATA transcription factor in breast cancer. Horm Cancer. 2011;2:132–43.
Sanchez-Garcia FL, Villagrasa P, Matsui J, Kotliar D, Castro VN, Akavia U-D, Chen B-J, Saucedo-Cuevas L, Rodriguez Barrueco R, Llobet-Navas D, Silva JM, Pe er D. Integration of genomic data enables selective discovery of breast cancer drivers. Cell. 2014;159:1461–75.
Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33.
Kurihara I, Lee D-K, Petit FG, Jeong J, Lee K, Lydon JP, DeMayo FJ, Tsai M-J, Tsai SY. COUP-TFII mediates progesterone regulation of uterine implantation by controlling ER activity. PLoS Genet. 2007;3:e102.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137.
Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–23.
Cerase A, Pintacuda G, Tattermusch A, Avner P. Xist localization and function: new insights from multiple levels. Genome Biol. 2015;16:166.
Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ScChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–8.
Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M, Carroll JS. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 2010;24:171–82.
Wang S, Sun H, Ma J, Zang C, Wang C, Wang J, Tang Q, Meyer CA, Zhang Y, Liu XS. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nature protocols. 2013;8:2502–2515.
Acknowledgements
We would like to thank Clive D’Santos and Chris Taylor from the CRUK CI Proteomics core, James Hadfield and the other members of the Genomics core, and Suraj Menon from the Bioinformatics core. Jason Carroll is supported by an ERC Consolidator grant and a Komen Scholar Award. We would like to acknowledge the support of the University of Cambridge, Cancer Research UK and Hutchison Whampoa Limited.
Author contributions
AAS, HM, KWM and JSC designed the project and wrote the manuscript. AAS, HM and AM performed the experiments and analysed the data.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
These authors contributed equally: A.A. Serandour and H. Mohammed.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Serandour, A.A., Mohammed, H., Miremadi, A. et al. TRPS1 regulates oestrogen receptor binding and histone acetylation at enhancers. Oncogene 37, 5281–5291 (2018). https://doi.org/10.1038/s41388-018-0312-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0312-2
This article is cited by
-
TRPS1 regulates the opposite effect of progesterone via RANKL in endometrial carcinoma and breast carcinoma
Cell Death Discovery (2023)