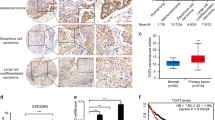
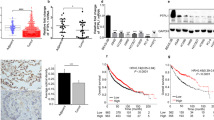
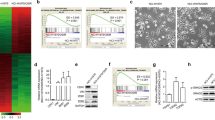
Abstract
Epithelial–mesenchymal transition (EMT) is an important process for cancer metastasis, drug resistance, and cancer stem cells. Activation of fibroblast growth factor receptor 1 (FGFR1) was found to promote EMT and metastasis in prostate and breast cancers, but the effects and mechanisms in lung cancer was unclear. In this study, we aimed to explore whether and how activation of FGFR1 promotes EMT and metastasis in FGFR1-amplified lung cancer. We show that activation of FGFR1 by its ligand fibroblast growth factor 2 (FGF2) promoted proliferation, EMT, migration, and invasion in FGFR1-amplified lung cancer cell lines H1581 and DMS114, whereas inhibition of FGFR1 suppressed these processes. FGFR1 activation upregulated expression of Sry-related HMG box 2 (SOX2) by downstream phosphorylated ERK1/2; moreover, the upregulation of SOX2 by autophosphorylation variant ERK2_R67S plasmid transfection was not suppressed by FGFR1 inhibitor AZD4547 or MEK/ERK inhibitor AZD6244 in vitro. And SOX2 expression was also significantly upregulated in ERK2_R67S lentivirus-transfected stable cell lines in vivo. Overexpression of SOX2 promoted cell proliferation, EMT, migration, and invasion. Importantly, activation of FGFR1 could not promote these processes in SOX2-silenced stable cell lines. In orthotopic and subcutaneous lung cancer xenograft models, inhibition of FGFR1 suppressed tumor growth, SOX2 expression, EMT, and metastasis in vivo; however, these processes caused by SOX2-overexpressing stable cell lines were not suppressed by FGFR1 inhibition. Higher expression of FGFR1 and SOX2 were positively correlated, and both were associated with shorter survival in lung cancer patients. In conclusion, our findings reveal that activation of FGFR1 promotes cell proliferation, EMT, and metastasis by the newly defined FGFR1-ERK1/2-SOX2 axis in FGFR1-amplified lung cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
07 September 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30.
Lewis DR, Check DP, Caporaso NE, Travis WD, Devesa SS. US lung cancer trends by histologic type. Cancer. 2014;120:2883–92.
Hanna N, Johnson D, Temin S, Baker S Jr., Brahmer J, Ellis PM, et al. Systemic therapy for stage IV non-small-cell lung cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol. 2017;35:3484–515.
Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25.
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50.
George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53.
The Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine (NGM). A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5:209ra153
Chen F, Zhang Y, Parra E, Rodriguez J, Behrens C, Akbani R, et al. Multiplatform-based molecular subtypes of non-small-cell lung cancer. Oncogene. 2017;36:1384–93.
Kim HR, Kim DJ, Kang DR, Lee JG, Lim SM, Lee CY, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol. 2013;31:731–7.
Sousa V, Reis D, Silva M, Alarcao AM, Ladeirinha AF, d’Aguiar MJ, et al. Amplification of FGFR1 gene and expression of FGFR1 protein is found in different histological types of lung carcinoma. Virchows Arch. 2016;469:173–82.
Schultheis AM, Bos M, Schmitz K, Wilsberg L, Binot E, Wolf J, et al. Fibroblast growth factor receptor 1 (FGFR1) amplification is a potential therapeutic target in small-cell lung cancer. Mod Pathol. 2014;27:214–21.
Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93.
Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS ONE. 2011;6:e20351.
Thomas A, Lee JH, Abdullaev Z, Park KS, Pineda M, Saidkhodjaeva L, et al. Characterization of fibroblast growth factor receptor 1 in small-cell lung cancer. J Thorac Oncol. 2014;9:567–71.
Malchers F, Dietlein F, Schottle J, Lu X, Nogova L, Albus K, et al. Cell-autonomous and non-cell-autonomous mechanisms of transformation by amplified FGFR1 in lung cancer. Cancer Discov. 2014;4:246–57.
Paik PK, Shen R, Berger MF, Ferry D, Soria JC, Mathewson A, et al. A phase Ib open-label multicenter study of AZD4547 in patients with advanced squamous cell lung cancers. Clin Cancer Res. 2017;23:5366–73.
Nogova L, Sequist LV, Perez Garcia JM, Andre F, Delord JP, Hidalgo M, et al. Evaluation of BGJ398, a fibroblast growth factor receptor 1-3 kinase inhibitor, in patients with advanced solid tumors harboring genetic alterations in fibroblast growth factor receptors: results of a global phase I, dose-escalation and dose-expansion study. J Clin Oncol. 2017;35:157–65.
Lim SH, Sun JM, Choi YL, Kim HR, Ahn S, Lee JY, et al. Efficacy and safety of dovitinib in pretreated patients with advanced squamous non-small cell lung cancer with FGFR1 amplification: a single-arm, phase 2 study. Cancer. 2016;122:3024–31.
Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8.
Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21–45.
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96.
De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110.
Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol. 2012;22:396–403.
Terry S, Savagner P, Ortiz-Cuaran S, Mahjoubi L, Saintigny P, Thiery JP, et al. New insights into the role of EMT in tumor immune escape. Mol Oncol. 2017;11:824–46.
Dieci MV, Arnedos M, Andre F, Soria JC. Fibroblast growth factor receptor inhibitors as a cancer treatment: from a biologic rationale to medical perspectives. Cancer Discov. 2013;3:264–79.
Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Med Res Rev. 2014;34:280–300.
Katoh M. Therapeutics targeting FGF signaling network in human diseases. Trends Pharmacol Sci. 2016;37:1081–96.
Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, et al. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell. 2007;12:559–71.
Brown WS, Tan L, Smith A, Gray NS, Wendt MK. Covalent targeting of fibroblast growth factor receptor inhibits metastatic breast cancer. Mol Cancer Ther. 2016;15:2096–106.
Jakobsen KR, Demuth C, Madsen AT, Hussmann D, Vad-Nielsen J, Nielsen AL, et al. MET amplification and epithelial-to-mesenchymal transition exist as parallel resistance mechanisms in erlotinib-resistant, EGFR-mutated, NSCLC HCC827 cells. Oncogenesis. 2017;6:e307.
Ji W, Yu Y, Li Z, Wang G, Li F, Xia W, et al. FGFR1 promotes the stem cell-like phenotype of FGFR1-amplified non-small cell lung cancer cells through the Hedgehog pathway. Oncotarget. 2016;7:15118–34.
Yuan H, Li ZM, Shao J, Ji WX, Xia W, Lu S. FGF2/FGFR1 regulates autophagy in FGFR1-amplified non-small cell lung cancer cells. J Exp Clin Cancer Res. 2017;36:72.
Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30.
Weina K, Utikal J. SOX2 and cancer: current research and its implications in the clinic. Clin Transl Med. 2014;3:19.
Wang L, Yang H, Lei Z, Zhao J, Chen Y, Chen P, et al. Repression of TIF1gamma by SOX2 promotes TGF-beta-induced epithelial-mesenchymal transition in non-small-cell lung cancer. Oncogene. 2016;35:867–77.
Kim M, Jang K, Miller P, Picon-Ruiz M, Yeasky TM, El-Ashry D, et al. VEGFA links self-renewal and metastasis by inducing Sox2 to repress miR-452, driving Slug. Oncogene. 2017;36:5199–211.
Yeh TC, Marsh V, Bernat BA, Ballard J, Colwell H, Evans RJ, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res. 2007;13:1576–83.
Brady DC, Crowe MS, Turski ML, Hobbs GA, Yao X, Chaikuad A, et al. Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature. 2014;509:492–6.
Tan Q, Li F, Wang G, Xia W, Li Z, Niu X, et al. Identification of FGF19 as a prognostic marker and potential driver gene of lung squamous cell carcinomas in Chinese smoking patients. Oncotarget. 2016;7:18394–402.
Singleton KR, Hinz TK, Kleczko EK, Marek LA, Kwak J, Harp T, et al. Kinome RNAi screens reveal synergistic targeting of MTOR and FGFR1 pathways for treatment of lung cancer and HNSCC. Cancer Res. 2015;75:4398–406.
Kotani H, Ebi H, Kitai H, Nanjo S, Kita K, Huynh TG, et al. Co-active receptor tyrosine kinases mitigate the effect of FGFR inhibitors in FGFR1-amplified lung cancers with low FGFR1 protein expression. Oncogene. 2016;35:3587–97.
Malchers F, Ercanoglu M, Schutte D, Castiglione R, Tischler V, Michels S, et al. Mechanisms of primary drug resistance in FGFR1-amplified lung cancer. Clin Cancer Res. 2017;23:5527–36.
Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–95.
Jayson GC, Kerbel R, Ellis LM, Harris AL. Antiangiogenic therapy in oncology: current status and future directions. Lancet. 2016;388:518–29.
Hu Y, Feng X, Mintz A, Jeffrey Petty W, Hsu W. Regulation of brachyury by fibroblast growth factor receptor 1 in lung cancer. Oncotarget. 2016;7:87124–35.
Maehara O, Suda G, Natsuizaka M, Ohnishi S, Komatsu Y, Sato F, et al. Fibroblast growth factor-2-mediated FGFR/Erk signaling supports maintenance of cancer stem-like cells in esophageal squamous cell carcinoma. Carcinogenesis. 2017;38:1073–83.
Chen Y, Gu H, Zhang DS, Li F, Liu T, Xia W. Highly effective inhibition of lung cancer growth and metastasis by systemic delivery of siRNA via multimodal mesoporous silica-based nanocarrier. Biomaterials. 2014;35:10058–69.
Peng L, Feng L, Yuan H, Benhabbour SR, Mumper RJ. Development of a novel orthotopic non-small cell lung cancer model and therapeutic benefit of 2ʹ-(2-bromohexadecanoyl)-docetaxel conjugate nanoparticles. Nanomedicine. 2014;10:1497–506.
Brewer JR, Mazot P, Soriano P.Genetic insights into the mechanisms of Fgf signaling.Genes Dev. 2016;30:751–71.
Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72:2045–56.
Zhang J, Zhang L, Su X, Li M, Xie L, Malchers F, et al. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res. 2012;18:6658–67.
Mandal R, Becker S, Strebhardt K. Stamping out RAF and MEK1/2 to inhibit the ERK1/2 pathway: an emerging threat to anticancer therapy. Oncogene. 2016;35:2547–61.
Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–50.
Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602.
Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–42.
Ferone G, Song JY, Sutherland KD, Bhaskaran R, Monkhorst K, Lambooij JP, et al. SOX2 is the determining oncogenic switch in promoting lung squamous cell carcinoma from different cells of origin. Cancer Cell. 2016;30:519–32.
Justilien V, Walsh MP, Ali SA, Thompson EA, Murray NR, Fields AP. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell. 2014;25:139–51.
Tripathi SC, Fahrmann JF, Celiktas M, Aguilar M, Marini KD, Jolly MK, et al. MCAM mediates chemoresistance in small-cell lung cancer via the PI3K/AKT/SOX2 signaling pathway. Cancer Res. 2017;77:4414–25.
Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–10.
Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, et al. Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell. 2016;165:45–60.
Taniguchi J, Pandian GN, Hidaka T, Hashiya K, Bando T, Kim KK, et al. A synthetic DNA-binding inhibitor of SOX2 guides human induced pluripotent stem cells to differentiate into mesoderm. Nucleic Acids Res. 2017;45:9219–28.
Datta J, Damodaran S, Parks H, Ocrainiciuc C, Miya J, Yu L, et al. Akt activation mediates acquired resistance to fibroblast growth factor receptor inhibitor BGJ398. Mol Cancer Ther. 2017;16:614–24.
Adachi Y, Watanabe K, Kita K, Kitai H, Kotani H, Sato Y, et al. Resistance mediated by alternative receptor tyrosine kinases in FGFR1-amplified lung cancer. Carcinogenesis. 2017;38:1063–72.
Kim HR, Kang HN, Shim HS, Kim EY, Kim J, Kim DJ, et al. Co-clinical trials demonstrate predictive biomarkers for dovitinib, an FGFR inhibitor, in lung squamous cell carcinoma. Ann Oncol. 2017;28:1250–9.
Goke F, Franzen A, Hinz TK, Marek LA, Yoon P, Sharma R, et al. FGFR1 expression levels predict BGJ398 sensitivity of FGFR1-dependent head and neck squamous cell cancers. Clin Cancer Res. 2015;21:4356–64.
Toschi L, Finocchiaro G, Nguyen TT, Skokan MC, Giordano L, Gianoncelli L, et al. Increased SOX2 gene copy number is associated with FGFR1 and PIK3CA gene gain in non-small cell lung cancer and predicts improved survival in early stage disease. PLoS ONE. 2014;9:e95303.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. 2013;8:e82241.
Acknowledgements
This work was funded by the National Key R&D Program of China (2016YFC1303300 to S.L.), the National Natural Science Foundation of China (81672272 to S.L. and 81773115 to W.X.), the National Key Grant of China (2016YFC0906400 to W.X.), the Key Project of Shanghai Health & Family Planning Commission (201540365 to S.L.), Shanghai Municipal Science & Technology Commission Research Project (17431906103 to S.L.), and Shanghai Scientific Research Projects (14140902800 to S.L.) We thank Wangxi Hai and Med-X Ruijin Hospital Micro PET/CT Research Center for the microCT scan and analysis. We thank AstraZeneca Pharmaceutical Company for kindly supply of AZD4547 and AstraZeneca Research Funding.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, K., Ji, W., Yu, Y. et al. FGFR1-ERK1/2-SOX2 axis promotes cell proliferation, epithelial–mesenchymal transition, and metastasis in FGFR1-amplified lung cancer. Oncogene 37, 5340–5354 (2018). https://doi.org/10.1038/s41388-018-0311-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0311-3
This article is cited by
-
circNINL facilitates aerobic glycolysis, proliferation, invasion, and migration in lung cancer by sponging miR-3918 to mediate FGFR1 expression
European Journal of Medical Research (2024)
-
SOX4/HDAC2 Axis Enhances Cell Survivability and Reduces Apoptosis by Activating AKT/MAPK Signaling in Colorectal Cancer
Digestive Diseases and Sciences (2024)
-
Targeting FGFRs by pemigatinib induces G1 phase cell cycle arrest, cellular stress and upregulation of tumor suppressor microRNAs
Journal of Translational Medicine (2023)
-
Rare molecular subtypes of lung cancer
Nature Reviews Clinical Oncology (2023)
-
GSK3β-driven SOX2 overexpression is a targetable vulnerability in esophageal squamous cell carcinoma
Oncogene (2023)