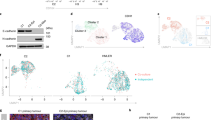
Abstract
An epithelial to mesenchymal transition (EMT) has been correlated to malignant tumor progression and metastasis by promoting cancer cell migration and invasion and chemoresistance. Hence, finding druggable EMT effectors is critical to efficiently interfere with metastasis formation and to overcome therapy resistance. We have employed a high-content microscopy screen in combination with a kinome and phosphatome-wide siRNA library to identify signaling pathways underlying an EMT of murine mammary epithelial cells and breast cancer cells. This screen identified the MEK5–ERK5 axis as a critical player in TGFβ-mediated EMT. Suppression of MEK5–ERK5 signaling completely prevented the morphological and molecular changes occurring during a TGFβ-induced EMT and, conversely, forced highly metastatic breast cancer cells into a differentiated epithelial state. Inhibition of MEK5–ERK5 signaling also repressed breast cancer cell migration and invasion and substantially reduced lung metastasis without affecting primary tumor growth. The results suggest that the MEK5–ERK5 signaling axis via activation of MEF2B and other transcription factors plays an important role in the induction and maintenance of breast cancer cell migration and invasion and thus represents an exploitable target for the pharmacological inhibition of cancer cell metastasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90.
Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850.
Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35:645–54.
Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8.
Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518–29.
Diepenbruck M, Waldmeier L, Ivanek R, Berninger P, Arnold P, van Nimwegen E, et al. Tead2 expression levels control the subcellular distribution of Yap and Taz, zyxin expression and epithelial-mesenchymal transition. J Cell Sci. 2014;127(Pt 7):1523–36.
Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768–83.
Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–30.
Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–6.
Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE. 2008;3:e2888.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell . 2008;133:704–15.
Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol. 2012;22:396–403.
Kalluri R. EMT: when epithelial cells decide to become mesenchymal-like cells. J Clin Invest. 2009;119:1417–9.
Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73.
Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–42.
Cavallaro U, Schaffhauser B, Christofori G. Cadherins and the tumour progression: is it all in a switch? Cancer Lett. 2002;176:123–8.
Tse JC, Kalluri R. Mechanisms of metastasis: epithelial-to-mesenchymal transition and contribution of tumor microenvironment. J Cell Biochem. 2007;101:816–29.
Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75:50–83.
Yan C, Luo H, Lee JD, Abe J, Berk BC. Molecular cloning of mouse ERK5/BMK1 splice variants and characterization of ERK5 functional domains. J Biol Chem. 2001;276:10870–8.
Kato Y, Zhao M, Morikawa A, Sugiyama T, Chakravortty D, Koide N, et al. Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J Biol Chem. 2000;275:18534–40.
English JM, Pearson G, Baer R, Cobb MH. Identification of substrates and regulators of the mitogen-activated protein kinase ERK5 using chimeric protein kinases. J Biol Chem. 1998;273:3854–60.
Zhou G, Bao ZQ, Dixon JE. Components of a new human protein kinase signal transduction pathway. J Biol Chem. 1995;270:12665–9.
Mody N, Campbell DG, Morrice N, Peggie M, Cohen P. An analysis of the phosphorylation and activation of extracellular-signal-regulated protein kinase 5 (ERK5) by mitogen-activated protein kinase kinase 5 (MKK5) in vitro. Biochem J. 2003;372(Pt 2):567–75.
Kamakura S, Moriguchi T, Nishida E. Activation of the protein kinase ERK5/BMK1 by receptor tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. J Biol Chem. 1999;274:26563–71.
Gui T, Sun Y, Shimokado A, Muragaki Y. The roles of mitogen-activated protein kinase pathways in TGF-beta-induced epithelial-mesenchymal transition. J Signal Transduct. 2012;2012:289243.
Marchetti A, Colletti M, Cozzolino AM, Steindler C, Lunadei M, Mancone C, et al. ERK5/MAPK is activated by TGFbeta in hepatocytes and required for the GSK-3beta-mediated Snail protein stabilization. Cell Signal. 2008;20:2113–8.
Badshah II, Baines DL, Dockrell ME. Erk5 is a mediator to TGFbeta1-induced loss of phenotype and function in human podocytes. Front Pharmacol. 2014;5:71.
Chen R, Yang Q, Lee JD. BMK1 kinase suppresses epithelial-mesenchymal transition through the Akt/GSK3beta signaling pathway. Cancer Res. 2012;72:1579–87.
Javaid S, Zhang J, Smolen GA, Yu M, Wittner BS, Singh A, et al. MAPK7 regulates EMT features and modulates the generation of CTCs. Mol Cancer Res. 2015;13:934–43.
Lotz-Jenne C, Luthi U, Ackerknecht S, Lehembre F, Fink T, Stritt M, et al. A high-content EMT screen identifies multiple receptor tyrosine kinase inhibitors with activity on TGFbeta receptor. Oncotarget. 2016;7:25983–6002.
Maeda M, Johnson KR, Wheelock MJ. Cadherin switching: essential for behavioral but not morphological changes during an epithelium-to-mesenchyme transition. J Cell Sci. 2005;118(Pt 5):873–87.
Zhang Y, Feng X, We R, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-beta response. Nature. 1996;383:168–72.
Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov. 2012;11:790–811.
Nakamura K, Johnson GL. PB1 domains of MEKK2 and MEKK3 interact with the MEK5 PB1 domain for activation of the ERK5 pathway. J Biol Chem. 2003;278:36989–92.
Chao TH, Hayashi M, Tapping RI, Kato Y, Lee JD. MEKK3 directly regulates MEK5 activity as part of the big mitogen-activated protein kinase 1 (BMK1) signaling pathway. J Biol Chem. 1999;274:36035–8.
Diaz-Meco MT, Moscat J. MEK5, a new target of the atypical protein kinase C isoforms in mitogenic signaling. Mol Cell Biol. 2001;21:1218–27.
Deacon K, Blank JL. Characterization of the mitogen-activated protein kinase kinase 4 (MKK4)/c-Jun NH2-terminal kinase 1 and MKK3/p38 pathways regulated by MEK kinases 2 and 3. MEK kinase 3 activates MKK3 but does not cause activation of p38 kinase in vivo. J Biol Chem. 1997;272:14489–96.
Whitmarsh AJ, Davis RJ. Role of mitogen-activated protein kinase kinase 4 in cancer. Oncogene. 2007;26:3172–84.
Sun W, Vincent S, Settleman J, Johnson GL. MEK kinase 2 binds and activates protein kinase C-related kinase 2. Bifurcation of kinase regulatory pathways at the level of an MAPK kinase kinase. J Biol Chem. 2000;275:24421–8.
Vincent S, Settleman J. The PRK2 kinase is a potential effector target of both Rho and Rac GTPases and regulates actin cytoskeletal organization. Mol Cell Biol. 1997;17:2247–56.
Campisi J, d’Adda, di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–40.
Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–405.
Lou Y, Preobrazhenska O, auf dem Keller U, Sutcliffe M, Barclay L, McDonald PC, et al. Epithelial-mesenchymal transition (EMT) is not sufficient for spontaneous murine breast cancer metastasis. Dev Dyn. 2008;237:2755–68.
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39.
Minn AJ, Kang Y, Serganova I, Gupta GP, Giri DD, Doubrovin M, et al. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest. 2005;115:44–55.
Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–5.
von Minckwitz G. Docetaxel/anthracycline combinations for breast cancer treatment. Expert Opin Pharmacother. 2007;8:485–95.
Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2016;44(D1):D481–7.
Milacic M, Haw R, Rothfels K, Wu G, Croft D, Hermjakob H, et al. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers. 2012;4:1180–211.
Balwierz PJ, Pachkov M, Arnold P, Gruber AJ, Zavolan M, van Nimwegen E. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 2014;24:869–84.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–7.
Kwon AT, Arenillas DJ, Worsley Hunt R, Wasserman WW. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3. 2012;2:987–1002.
Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol. 2016;43:7–13.
Akhurst RJ, Derynck R. TGF-beta signaling in cancer--a double-edged sword. Trends Cell Biol. 2001;11:S44–51.
Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–7.
Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–8.
Yang Q, Deng X, Lu B, Cameron M, Fearns C, Patricelli MP, et al. Pharmacological inhibition of BMK1 suppresses tumor growth through promyelocytic leukemia protein. Cancer Cell. 2010;18:258–67.
El-Deiry WS. p21(WAF1) mediates cell-cycle inhibition, relevant to cancer suppression and therapy. Cancer Res. 2016;76:5189–91.
Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–46.
Klein AM, Zaganjor E, Cobb MH. Chromatin-tethered MAPKs. Curr Opin Cell Biol. 2013;25:272–7.
Turjanski AG, Vaque JP, Gutkind JS. MAP kinases and the control of nuclear events. Oncogene. 2007;26:3240–53.
Hodge RG, Ridley AJ. Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol. 2016;17:496–510.
Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907.
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601.
Laurent J, Hull EF, Touvrey C, Kuonen F, Lan Q, Lorusso G, et al. Proangiogenic factor PlGF programs CD11b(+) myelomonocytes in breast cancer during differentiation of their hematopoietic progenitors. Cancer Res. 2011;71:3781–91.
Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73.
Acknowledgements
We thank P. Lorentz and the DBM microscopy facility (DBM, University of Basel) for excellent support with microscopy, H. Antoniadis, P. Schmidt, and I. Galm for technical support, and E. Fagiani for flow cytometry. We also thank the microscopy facility at Actelion Pharmaceutical, in particular Carina Lotz-Jenne, for input to the screening experiments, and F. Tang for help with phostag gels.
Funding
This work was supported by the SystemsX.ch MTD project MetastasiX, the Swiss National Science Foundation, the Swiss Cancer League, and Krebsliga Beider Basel. N.M.-S. was supported by a Marie-Heim Vögtlin grant from the Swiss National Foundation.
Author contributions
S.P. designed and performed the experiments, analyzed the data and wrote the paper. N.M.-S. helped in the screening set up, performed experiments and analyzed the data. M.D. performed experiments and analyzed data. M.S. isolated primary tumor cells. R.K.R.K. performed bioinformatics analysis. G.C. oversaw the project, designed experiments, analyzed data and wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
These authors contributed equally: Nathalie Meyer-Schaller, Maren Diepenbruck.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Pavan, S., Meyer-Schaller, N., Diepenbruck, M. et al. A kinome-wide high-content siRNA screen identifies MEK5–ERK5 signaling as critical for breast cancer cell EMT and metastasis. Oncogene 37, 4197–4213 (2018). https://doi.org/10.1038/s41388-018-0270-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0270-8
This article is cited by
-
The ERK5/NF-κB signaling pathway targets endometrial cancer proliferation and survival
Cellular and Molecular Life Sciences (2022)
-
The extracellular-regulated protein kinase 5 (ERK5) enhances metastatic burden in triple-negative breast cancer through focal adhesion protein kinase (FAK)-mediated regulation of cell adhesion
Oncogene (2021)
-
RETRACTED ARTICLE: LncRNA XIST inhibits ovarian cancer cell growth and metastasis via regulating miR-150-5p/PDCD4 signaling pathway
Naunyn-Schmiedeberg's Archives of Pharmacology (2021)
-
A dual role of Irf1 in maintaining epithelial identity but also enabling EMT and metastasis formation of breast cancer cells
Oncogene (2020)
-
Targeting MEK5 impairs nonhomologous end-joining repair and sensitizes prostate cancer to DNA damaging agents
Oncogene (2020)