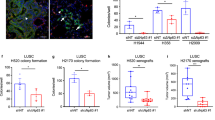
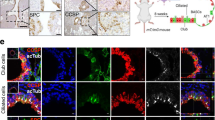
Abstract
HOXB7 is a homeodomain (HOX) transcription factor involved in regional body patterning of invertebrates and vertebrates. We previously identified HOXB7 within a ten-gene prognostic signature for lung adenocarcinoma, where increased expression of HOXB7 was associated with poor prognosis. This raises the question of how HOXB7 overexpression can influence the metastatic behavior of lung adenocarcinoma. Here, we analyzed publicly available microarray and RNA-seq lung cancer expression datasets and found that HOXB7-overexpressing tumors are enriched in gene signatures characterizing adult and embryonic stem cells (SC), and induced pluripotent stem cells (iPSC). Experimentally, we found that HOXB7 upregulates several canonical SC/iPSC markers and sustains the expansion of a subpopulation of cells with SC characteristics, through modulation of LIN28B, an emerging cancer gene and pluripotency factor, which we discovered to be a direct target of HOXB7. We validated this new circuit by showing that HOXB7 enhances reprogramming to iPSC with comparable efficiency to LIN28B or its target c-MYC, which is a canonical reprogramming factor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801.
Riddihough G. Developmental biology. Homing in on the homeobox. Nature. 1992;357:643–4.
Kornberg TB. Understanding the homeodomain. J Biol Chem. 1993;268:26813–6.
Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–76.
Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904.
Morgan R. Hox genes: a continuation of embryonic patterning? Trends Genet. 2006;22:67–69.
Care A, Silvani A, Meccia E, Mattia G, Stoppacciaro A, Parmiani G, et al. HOXB7 constitutively activates basic fibroblast growth factor in melanomas. Mol Cell Biol. 1996;16:4842–51.
Storti P, Donofrio G, Colla S, Airoldi I, Bolzoni M, Agnelli L, et al. HOXB7 expression by myeloma cells regulates their pro-angiogenic properties in multiple myeloma patients. Leukemia. 2011;25:527–37.
Care A, Silvani A, Meccia E, Mattia G, Peschle C, Colombo MP. Transduction of the SkBr3 breast carcinoma cell line with the HOXB7 gene induces bFGF expression, increases cell proliferation and reduces growth factor dependence. Oncogene. 1998;16:3285–9.
Wu X, Chen H, Parker B, Rubin E, Zhu T, Lee JS, et al. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66:9527–34.
Jin K, Kong X, Shah T, Penet MF, Wildes F, Sgroi DC, et al. The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proc Natl Acad Sci USA. 2012;109:2736–41.
Liao WT, Jiang D, Yuan J, Cui YM, Shi XW, Chen CM, et al. HOXB7 as a prognostic factor and mediator of colorectal cancer progression. Clin Cancer Res: Off J Am Assoc Cancer Res. 2011;17:3569–78.
Nguyen Kovochich A, Arensman M, Lay AR, Rao NP, Donahue T, Li X, et al. HOXB7 promotes invasion and predicts survival in pancreatic adenocarcinoma. Cancer. 2013;119:529–39.
De Souza Setubal Destro MF, Bitu CC, Zecchin KG, Graner E, Lopes MA, Kowalski LP, et al. Overexpression of HOXB7 homeobox gene in oral cancer induces cellular proliferation and is associated with poor prognosis. Int J Oncol. 2010;36:141–9.
Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20.
Bianchi F, Nuciforo P, Vecchi M, Bernard L, Tizzoni L, Marchetti A, et al. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. J Clin Invest. 2007;117:3436–44.
Dama E, Melocchi V, Dezi F, Pirroni S, Carletti RM, Brambilla D, et al. An aggressive subtype of Stage I lung adenocarcinoma with molecular and prognostic characteristics typical of advanced lung cancers. Clin Cancer Res: Off J Am Assoc Cancer Res. 2017;23:62–72.
Yuan W, Zhang X, Xu Y, Li S, Hu Y, Wu S. Role of HOXB7 in regulation of progression and metastasis of human lung adenocarcinoma. Mol Carcinog. 2014;53:49–57.
Errico MC, Jin K, Sukumar S, Care A. The widening sphere of influence of HOXB7 in solid tumors. Cancer Res. 2016;76:2857–62.
Liu S, Jin K, Hui Y, Fu J, Jie C, Feng S, et al. HOXB7 promotes malignant progression by activating the TGFbeta signaling pathway. Cancer Res. 2015;75:709–19.
Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50.
Director’s Challenge Consortium for the Molecular Classification of Lung A, Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–7.
Yamauchi M, Yamaguchi R, Nakata A, Kohno T, Nagasaki M, Shimamura T, et al. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PLoS ONE. 2012;7:e43923.
Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70.
Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–14.
Wang P, Gao Q, Suo Z, Munthe E, Solberg S, Ma L, et al. Identification and characterization of cells with cancer stem cell properties in human primary lung cancer cell lines. PLoS ONE. 2013;8:e57020.
Lu H, Clauser KR, Tam WL, Frose J, Ye X, Eaton EN, et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat Cell Biol. 2014;16:1105–17.
Yan X, Luo H, Zhou X, Zhu B, Wang Y, Bian X. Identification of CD90 as a marker for lung cancer stem cells in A549 and H446 cell lines. Oncol Rep. 2013;30:2733–40.
Whitfield TW, Wang J, Collins PJ, Partridge EC, Aldred SF, Trinklein ND, et al. Functional analysis of transcription factor binding sites in human promoters. Genome Biol. 2012;13:R50.
Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci USA. 2009;106:3384–9.
Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–49.
Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706.
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47.
Liu Y, Li H, Feng J, Cui X, Huang W, Li Y, et al. Lin28 induces epithelial-to-mesenchymal transition and stemness via downregulation of let-7a in breast cancer cells. PLoS ONE. 2013;8:e83083.
Copley MR, Babovic S, Benz C, Knapp DJ, Beer PA, Kent DG, et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15:916–25.
Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–9.
Shao Y, Zhang L, Cui L, Lou W, Wang D, Lu W, et al. LIN28B suppresses microRNA let-7b expression to promote CD44+/LIN28B+ human pancreatic cancer stem cell proliferation and invasion. Am J Cancer Res. 2015;5:2643–59.
Chien CS, Wang ML, Chu PY, Chang YL, Liu WH, Yu CC, et al. Lin28B/Let-7 regulates expression of Oct4 and Sox2 and reprograms oral squamous cell carcinoma cells to a stem-like state. Cancer Res. 2015;75:2553–65.
Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–8.
Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–70.
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76.
Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12.
Yamanaka S, Tanabe K. Method of efficiently establishing induced pluripotent stem cells. Google Patents, 2011; US 20160122720 A1.
Hammachi F, Morrison GM, Sharov AA, Livigni A, Narayan S, Papapetrou EP, et al. Transcriptional activation by Oct4 is sufficient for the maintenance and induction of pluripotency. Cell Rep. 2012;1:99–109.
Wesselschmidt RL. The teratoma assay: an in vivo assessment of pluripotency. Methods Mol Biol. 2011;767:231–41.
Huan HB, Yang DP, Wen XD, Chen XJ, Zhang L, Wu LL, et al. HOXB7 accelerates the malignant progression of hepatocellular carcinoma by promoting stemness and epithelial−mesenchymal transition. J Exp Clin Cancer Res. 2017;36:86.
Allavena P, Sica A, Solinas G, Porta C, Mantovani A. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Crit Rev Oncol Hematol. 2008;66:1–9.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial−mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15.
Wetzel A, Chavakis T, Preissner KT, Sticherling M, Haustein UF, Anderegg U, et al. Human Thy-1 (CD90) on activated endothelial cells is a counterreceptor for the leukocyte integrin Mac-1 (CD11b/CD18). J Immunol. 2004;172:3850–9.
Wang X, Cao L, Wang Y, Wang X, Liu N, You Y. Regulation of let-7 and its target oncogenes (Review). Oncol Lett. 2012;3:955–60.
Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–91.
He X, Liu Z, Xia Y, Xu J, Lv G, Wang L, et al. HOXB7 overexpression promotes cell proliferation and correlates with poor prognosis in gastric cancer patients by inducing expression of both AKT and MARKs. Oncotarget. 2017;8:1247–61.
Care A, Valtieri M, Mattia G, Meccia E, Masella B, Luchetti L, et al. Enforced expression of HOXB7 promotes hematopoietic stem cell proliferation and myeloid-restricted progenitor differentiation. Oncogene. 1999;18:1993–2001.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010;7:53–55.
Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, et al. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–15.
Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–64.
Acknowledgements
We thank Bruno Amati for kindly providing the pBABE-MYC plasmid. We are indebted to Giuseppina Bonizzi, Giovanna Jodice, Elena Belloni, Elena Marino, Francesca De Santis, Stefania Pirroni, Pietro Vella, Valentina Melocchi, Davide Disalvatore and Stefano Freddi for technical support. We thank Ugo Cavallaro for inspiring discussions and comments on the paper. This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC) (MFAG17568 to FB; IG 10349 and 14040 and MCO 10.000 to PPDF) and from the Monzino Foundation (to PPDF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Monterisi, S., Lo Riso, P., Russo, K. et al. HOXB7 overexpression in lung cancer is a hallmark of acquired stem-like phenotype. Oncogene 37, 3575–3588 (2018). https://doi.org/10.1038/s41388-018-0229-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0229-9
This article is cited by
-
An organoid library of salivary gland tumors reveals subtype-specific characteristics and biomarkers
Journal of Experimental & Clinical Cancer Research (2022)
-
HOXB7 acts as an oncogenic biomarker in head and neck squamous cell carcinoma
Cancer Cell International (2021)
-
Aggressive early-stage lung adenocarcinoma is characterized by epithelial cell plasticity with acquirement of stem-like traits and immune evasion phenotype
Oncogene (2021)
-
Multiple roles of HOX proteins in Metastasis: Let me count the ways
Cancer and Metastasis Reviews (2020)
-
LncRNA HOXA-AS3 confers cisplatin resistance by interacting with HOXA3 in non-small-cell lung carcinoma cells
Oncogenesis (2019)