Abstract
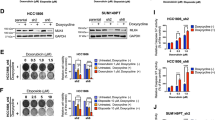
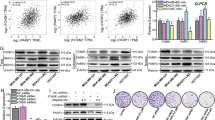
Liver receptor homolog-1 (LRH1) has been shown to promote tumor proliferation and development. However, the functions of LRH1 in mediating cancer cells chemoresistance are still not clear. Here, we found LRH1 levels were significantly elevated in primary breast cancer tissues in patients who developed early recurrence. Similarly, adriamycin (ADR)-resistant breast cancer cell lines also exerted high LRH1 expression. Indeed, overexpression of LRH1 attenuated cytotoxicity of chemotherapeutic drugs ADR and cisplatin (DDP) in breast cancer cells in vitro and in nude mice tumor model. Comet and BrdU assays showed overexpression of LRH1 blocked breast cancer cells DNA damage by chemotherapeutic drug, whereas depletion of LRH1 enhanced DNA damage. Remarkably, knockdown of LRH1 decreased the levels and foci of DNA damage marker γH2AX induced by ADR and DDP. Furthermore, plasmid end-joining assay indicated that knockdown of LRH1 significantly decreased non-homologous end-joining (NHEJ)-mediated double-strand break (DSB) repair efficiencies. Afterwards, we provided evidences that LRH1 promoted MDC1 transcription by directly activating MDC1 promoter and therefore increased γH2AX levels. Importantly, a LRH1-binding site mapped between −1812 and −1804 bp of the proximal MDC1 promoter was identified. Moreover, LRH1 and MDC1 mRNA levels were positively correlated in recurrent breast cancer samples. These results implied LRH1 enhanced breast cancer cell chemoresistance by upregulating MDC1 and attenuating DNA damage. Additionally, we elucidated the coactivator NCOA3 acted synergistically with LRH1 to promote MDC1 expression and chemoresistance. Altogether, LRH1-MDC1 signaling might be considered as a novel molecular target for designing novel therapeutic regimen in chemotherapy resistance breast cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–12.
Gilbert LA, Hemann MT. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143:355–66.
Gong C, Liu B, Yao Y, Qu S, Luo W, Tan W, et al. Potentiated DNA damage response in circulating breast tumor cells confers resistance to chemotherapy. J Biol Chem. 2015;290:14811–25.
Cree IA, Charlton P. Molecular chess? Hallmarks of anti-cancer drug resistance. BMC Cancer. 2017;17:10.
Ashwell S, Zabludoff S. DNA damage detection and repair pathways—recent advances with inhibitors of checkpoint kinases in cancer therapy. Clin Cancer Res. 2008;14:4032–7.
Fayard E, Auwerx J, Schoonjans K. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–60.
Gu PL, Goodwin B, Chung ACK, Xu XP, Wheeler DA, Price RR, et al. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol. 2005;25:3492–505.
Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–74.
Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–26.
Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–15.
Sirianni R, Seely JB, Attia G, Stocco DM, Carr BR, Pezzi V, et al. Liver receptor homologue-1 is expressed in human steroidogenic tissues and activates transcription of genes encoding steroidogenic enzymes. J Endocrinol. 2002;174:R13–7.
Benod C, Vinogradova MV, Jouravel N, Kim GE, Fletterick RJ, Sablin EP. Nuclear receptor liver receptor homologue 1 (LRH-1) regulates pancreatic cancer cell growth and proliferation. Proc Natl Acad Sci USA. 2011;108:16927–31.
Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–8.
Holmstrom SR, Deering T, Swift GH, Poelwijk FJ, Mangelsdorf DJ, Kliewer SA, et al. LRH-1 and PTF1-L coregulate an exocrine pancreas-specific transcriptional network for digestive function. Genes Dev. 2011;25:1674–9.
Schoonjans K, Dubuquoy L, Mebis J, Fayard E, Wendling O, Haby C, et al. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proc Natl Acad Sci USA. 2005;102:2058–62.
Kramer HB, Lai CF, Patel H, Periyasamy M, Lin ML, Feller SM, et al. LRH-1 drives colon cancer cell growth by repressing the expression of the CDKN1A gene in a p53-dependent manner. Nucleic Acids Res. 2016;44:582–94.
Sidler D, Renzulli P, Schnoz C, Berger B, Schneider-Jakob S, Fluck C, et al. Colon cancer cells produce immunoregulatory glucocorticoids. Oncogene. 2011;30:2411–9.
Bianco S, Jangal M, Garneau D, Gevry N. LRH-1 controls proliferation in breast tumor cells by regulating CDKN1A gene expression. Oncogene. 2015;34:4509–18.
Chand AL, Herridge KA, Thompson EW, Clyne CD. The orphan nuclear receptor LRH-1 promotes breast cancer motility and invasion. Endocr Relat Cancer. 2010;17:965–75.
Bianco S, Brunelle M, Jangal M, Magnani L, Gevry N. LRH-1 governs vital transcriptional programs in endocrine-sensitive and -resistant breast cancer cells. Cancer Res. 2014;74:2015–25.
Coster G, Goldberg M. The cellular response to DNA damage: a focus on MDC1 and its interacting proteins. Nucleus. 2010;1:166–78.
Goldberg M, Stucki M, Falck J, D’Amours D, Rahman D, Pappin D, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–6.
Stewart GS, Wang B, Bignell CR, Taylor AMR, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–6.
Jungmichel S, Clapperton JA, Lloyd J, Hari FJ, Spycher C, Pavic L, et al. The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator. Nucleic Acids Res. 2012;40:3913–28.
Liu J, Luo S, Zhao H, Liao J, Li J, Yang C, et al. Structural mechanism of the phosphorylation-dependent dimerization of the MDC1 forkhead-associated domain. Nucleic Acids Res. 2012;40:3898–912.
Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–26.
Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, et al. MDC1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–83.
Liu X, Dong R, Jiang Z, Wei Y, Li Y, Wei L, et al. MDC1 promotes ovarian cancer metastasis by inducing epithelial-mesenchymal transition. Tumour Biol. 2015;36:4261–9.
Luo X, Yao J, Nie P, Yang Z, Feng H, Chen P, et al. FOXM1 promotes invasion and migration of colorectal cancer cells partially dependent on HSPA5 transactivation. Oncotarget. 2016;7:26480–95.
Audet-Walsh E, Papadopoli DJ, Gravel SP, Yee T, Bridon G, Caron M, et al. The PGC-1alpha/ERRalpha axis represses one-carbon metabolism and promotes sensitivity to anti-folate therapy in breast cancer. Cell Rep. 2016;14:920–31.
Ao X, Nie P, Wu B, Xu W, Zhang T, Wang S, et al. Decreased expression of microRNA-17 and microRNA-20b promotes breast cancer resistance to taxol therapy by upregulation of NCOA3. Cell Death Dis. 2016;7:e2463.
Burgess RC, Burman B, Kruhlak MJ, Misteli T. Activation of DNA damage response signaling by condensed chromatin. Cell Rep. 2014;9:1703–17.
Baldock RA, Day M, Wilkinson OJ, Cloney R, Jeggo PA, Oliver AW, et al. ATM localization and heterochromatin repair depend on direct interaction of the 53BP1-BRCT2 domain with gammaH2AX. Cell Rep. 2015;13:2081–9.
Botrugno OA, Fayard E, Annicotte JS, Haby C, Brennan T, Wendling O, et al. Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Mol Cell. 2004;15:499–509.
Zhu J, Zou Z, Nie P, Kou X, Wu B, Wang S, et al. Downregulation of microRNA-27b-3p enhances tamoxifen resistance in breast cancer by increasing NR5A2 and CREB1 expression. Cell Death Dis. 2016;7:e2454.
Annicotte JS, Chavey C, Servant N, Teyssier J, Bardin A, Licznar A, et al. The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene. 2005;24:8167–75.
Thiruchelvam PT, Lai CF, Hua H, Thomas RS, Hurtado A, Hudson W, et al. The liver receptor homolog-1 regulates estrogen receptor expression in breast cancer cells. Breast Cancer Res Treat. 2011;127:385–96.
Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer. 2012;12:587–98.
Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–U651.
Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–11.
Gruosso T, Mieulet V, Cardon M, Bourachot B, Kieffer Y, Devun F, et al. Chronic oxidative stress promotes H2AX protein degradation and enhances chemosensitivity in breast cancer patients. EMBO Mol Med. 2016;8:527–49.
Lazarus KA, Wijayakumara D, Chand AL, Simpson ER, Clyne CD. Therapeutic potential of liver receptor homolog-1 modulators. J Steroid Biochem Mol Biol. 2012;130:138–46.
Burwinkel B, Wirtenberger M, Klaes R, Schmutzler RK, Grzybowska E, Forsti A, et al. Association of NCOA3 polymorphisms with breast cancer risk. Clin Cancer Res. 2005;11:2169–74.
Zou ZZ, Luo XY, Nie PP, Wu BY, Zhang T, Wei YC, et al. Inhibition of SRC-3 enhances sensitivity of human cancer cells to histone deacetylase inhibitors. Biochem Biophys Res Commun. 2016;478:227–33.
Lai CF, Flach KD, Alexi X, Fox SP, Ottaviani S, Thiruchelvam PT, et al. Co-regulated gene expression by oestrogen receptor alpha and liver receptor homolog-1 is a feature of the oestrogen response in breast cancer cells. Nucleic Acids Res. 2013;41:10228–40.
Coster G, Hayouka Z, Argaman L, Strauss C, Friedler A, Brandeis M, et al. The DNA damage response mediator MDC1 directly interacts with the anaphase-promoting complex/cyclosome. J Biol Chem. 2007;282:32053–64.
Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci USA. 2004;101:7624–9.
Zou ZZ, Yuan ZY, Zhang QX, Long ZJ, Chen JN, Tang ZP, et al. Aurora kinase A inhibition-induced autophagy triggers drug resistance in breast cancer cells. Autophagy. 2012;8:1798–810.
Zhang ZZ, Liu L, Jiang XX, Zhai SD, Xing D. The essential role of Drp1 and its regulation by S-nitrosylation of parkin in dopaminergic neurodegeneration: implications for Parkinson’s disease. Antioxid Redox Signal. 2016;25:609–22.
Zhang ZZ, Liu L, Wu SN, Xing D. Drp1, Mff, Fis1, and MiD51 are coordinated to mediate mitochondrial fission during UV irradiation-induced apoptosis. FASEB J. 2016;30:466–76.
Klammer H, Kadhim M, Iliakis G. Evidence of an adaptive response targeting DNA nonhomologous end joining and its transmission to bystander cells. Cancer Res. 2010;70:8498–506.
Acknowledgements
We are grateful to Dr. Simak Ali (Imperial College London, London) for providing pCI-HA3-LRH1 plasmid, to Dr. Michal Goldberg (Hebrew University of Jerusalem, Jerusalem) for providing HA-MDC1 plasmid, to Dr. Vera Gorbunova for providing the pEGFP-Pem1-Ad2 plasmid, to Prof. B-A Li (Xiamen University) for providing the MDA-MB-231/ADR and T47D/ADR cells. This study was supported by grants from the National Natural Science Foundation of China (No. 81772803, 81630046, 81402187); Ph.D. Start-Up Fund of Natural Science Foundation of Guangdong Province (No. 2014A030310505 to ZZ); Foundation for Distinguished Young Talents in Higher Education of Guangdong (No. C1085229 to ZZ).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
These authors contributed equally: S. Wang, Z. Zou.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Wang, S., Zou, Z., Luo, X. et al. LRH1 enhances cell resistance to chemotherapy by transcriptionally activating MDC1 expression and attenuating DNA damage in human breast cancer. Oncogene 37, 3243–3259 (2018). https://doi.org/10.1038/s41388-018-0193-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0193-4
This article is cited by
-
Photobiomodulation therapy moderates cancer cachexia-associated muscle wasting through activating PI3K/AKT/FoxO3a pathway
Apoptosis (2024)
-
LINC00472 inhibits cell migration by enhancing intercellular adhesion and regulates H3K27ac level via interacting with P300 in renal clear cell carcinoma
Cell Death Discovery (2022)
-
Macrophages and cancer stem cells: a malevolent alliance
Molecular Medicine (2021)
-
Prognostic significance and targeting tumor-associated macrophages in cancer: new insights and future perspectives
Breast Cancer (2021)
-
Prediction of blood-based biomarkers and subsequent design of bisulfite PCR-LDR-qPCR assay for breast cancer detection
BMC Cancer (2020)