Abstract
Using whole genome sequencing, we identified gene amplification of CREPT in colorectal cancer (CRC). In this study, we aim to clarify its clinical significance, biological effects, and mechanism in CRC. CREPT was upregulated in CRC cell lines and in 47.37% (72/152) of primary CRC tumors. Amplification of CREPT was detected in 48.28% (56/116) of primary CRC tumors, which was positively correlated with its overexpression (P < 0.001). Multivariate analysis showed that CRC patients with CREPT protein overexpression were significantly associated with poor disease-free survival (P < 0.05). CREPT significantly accelerated CRC cell proliferation and metastasis both in vitro and in vivo. RNA-sequencing (seq) analysis uncovered that the tumor-promoting effect by CREPT was attributed to enhancing Wnt/β-catenin signaling. Using co-immunoprecipitation coupled with mass spectroscopy, we identified p300 protein was a novel CREPT interacting partner. CREPT greatly increased the interaction between p300 and β-catenin, thus promoting p300-mediated β-catenin acetylation and stabilization. Moreover, CREPT cooperated with p300, leading to elevated active histone acetylation markers H3K27ac and H4Ac and decreased repressive histone marker H3K9me3 at the promoters of Wnt downstream targets. In summary, CREPT plays a pivotal oncogenic role in colorectal carcinogenesis through promoting Wnt/β-catenin pathway via cooperating with p300. CREPT may serve as a prognostic biomarker of patients with CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the third most common diagnosed cancer in men and the second in women worldwide [1]. More than 1 million individuals are diagnosed with CRC annually, with around 700,000 deaths occurring [2]. CRC is a multistep process with the accumulation of genetic and epigenetic alterations [3, 4]. Previous studies showed that somatic copy number alterations (CNAs) is crucial for the development of CRC [5]. Increased frequency of 7pq, 8q, 13q, and 20q gains, and of 4pq, 5q, 8p, 15q, 17p, and 18q losses were observed in colorectal adenoma and carcinoma progression [6,7,8,9]. Therefore, it is important to identify novel genes with CNAs and their biological functions in colorectal tumorigenesis.
Cell-cycle related and expression-elevated protein (CREPT) was firstly identified by our group as a potential oncogene [10]. The expression of CREPT was deregulated in many types of tumors [11,12,13,14,15]. CREPT is localized at human chromosome 20q11.23, which is a highly amplified region in CRC [16, 17]. CREPT functions as a scaffolds of C-terminal domain of human RNA polymerase II (RNAPII) and cooperates with RPAP2 to dephosphorylate of phospho-Ser5, thereby promoting gene transcription [18, 19]. We found that CREPT interacted and coordinated with RNAPII and β-catenin proteins, promoting oncogenic Wnt/β-catenin signaling and tumorigenesis [10, 20]. However, the role and the molecular mechanism of CREPT in CRC are still unclear. In this study, we investigated the expression regulation, clinical application, biological function, and molecular basis of CREPT in CRC.
Results
CREPT is upregulated in colon cancer cell lines and in primary CRC tissues
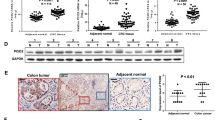
We first evaluated the expression of CREPT in CRC cell lines and human tissue samples. CREPT mRNA and protein expression was dramatically enhanced in all nine CRC cell lines compared with normal human colon tissues and two normal human colonic epithelial cells HCEC 1CT and 2CT [21] by real-time PCR and western blot (Fig. 1a, S Fig. 1a). Keeping with the findings, CREPT protein was significantly upregulated in 13 primary CRC tissues compared with their adjacent normal tissues by western blot (P = 0.002) (Fig. 1b). To further confirm our findings, the mRNA expression of CREPT in CRC samples from TCGA database (https://genome-cancer.ucsc.edu) was analyzed. CREPT was significantly increased in 32 CRC tissues compared with paired adjacent normal tissues (P < 0.0001) (Fig. 1c). The upregulation was also observed in unpaired CRC (n = 364) compared with normal samples (n = 50) from TCGA cohort (P < 0.0001) (Fig. 1d). Taken together, CREPT is significantly upregulated in CRC, indicating that it might play an oncogenic role in CRC.
CREPT was overexpressed in colon cancer cell lines and primary tissues. a The mRNA and protein level of CREPT was upregulated in most of the colon cancer cell lines detected by real-time PCR or western blot respectively. b The protein expression level of CREPT was significantly higher in primary CRC tumors as compared with their adjacent tissues by western blot. c The mRNA level of CREPT in tumors was dramatically elevated compared with their paired adjacent normal tissues from TCGA CRC database. d CREPT mRNA level was significantly increased in tumor tissues compared with normal tissues from TCGA CRC database (P < 0.0001). e Representative immunohistochemistry image of CREPT protein expression in CRC tumor tissues and their adjacent tissues. f Statistics histogram based on H-score of tissue microarray (n = 152) and normal tissues (n = 14) (P < 0.0001). g Disease-free survival in relation to expression was evaluated by the Kaplan–Meier survival curve and the log-rank test (P < 0.05) (g1). (g2) Patients at TNM I and II was defined as early stage
CREPT is an independent predictor of cancer recurrence in CRC patients
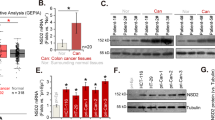
To evaluate the association of CREPT with the clinicopathological features and clinical outcomes of patients with CRC, we evaluated the CREPT protein expression in a tissue microarray of 152 CRC tumors. We found that 47.37% (72/152) of CRC tumors showed strong nuclear staining of CREPT, 29.61% (45/152) were moderate and 17.76% (27/152) were weak staining, and 5.26% (8/152) cases showed negative staining (Fig. 1e). CREPT was significantly upregulated in 152 tumor tissues compared with 14 normal colon tissues (P < 0.0001) (Fig. 1f). There was no significant correlation between CREPT expression and age, gender, tumor localization, differentiation, and the TNM stage of CRC patients (S Table 1). Kaplan–Meier analysis showed that CREPT high expression was significantly associated with poor disease-free survival in patients with CRC (P < 0.05, log-rank test) (Fig. 1g1, S Fig. 1b). Upregulated CREPT expression indicated a shorter disease-free survival of patients at early stage (TNM stages I and II, Fig. 1g2), but not at advanced stage (TNM stages III and IV, S Fig. 1b). In univariate Cox regression analysis, CREPT high expression was associated with an increased risk of cancer recurrence (RR = 1.897, 95%CI, 1.053−3.418; P < 0.05) (Table 1). After adjustment for potential confounding factors, multivariate Cox regression analysis showed that CREPT high expression was an independent predictor of cancer recurrence in patients with CRC (RR 1.897, 95% CI, 1.053−418; P < 0.05) (Table 1)
Copy number gain of CREPT contributes to its upregulation in CRC
Human CREPT gene localizes at the region q11.23 of chromosome 20, which is a highly amplified region in CRC [16, 17]. By whole genome sequencing analyses of the paired CRC tissues from our group, we identified CREPT amplification in CRC [17]. To further confirm our identification, we evaluated the copy number variation from TCGA (http://www.cbioportal.org/) and found the region that CREPT localizes was markedly amplified in both rectum cancer and colon cancer (Fig. 2a). We next examined the DNA copy number variation of CREPT in a cohort of 116 primary CRC tumor tissues using a specific Taqman probe against CREPT. CREPT copy number gain or amplification (defined as more than two copies) was found in 48.28% (56/116) CRC cases (Fig. 2b). Furthermore, a significantly positive correlation between CREPT mRNA level and its DNA copy number gain/amplification was observed using linear regression analysis (R = 0.458, P < 0.01) (Fig. 2c). Therefore, these results suggested that copy number gain of CREPT contributes to its upregulation in CRC.
CREPT gene was amplified in CRC. a Significant genomic alterations of chromosome 20p13-q13 across 12 while blue color refers to copy number loss. Red box indicates the location of CREPT gene. BLCA bladder urothelial carcinoma, BRCA breast invasive carcinoma, KIRC kidney renal clear cell carcinoma, COAD colorectal adenocarcinoma, GBM glioblastoma multiformed, HNSC head and neck squamous cell carcinoma, LAML acute myeloid leukemia, LUAD lung adenocarcinoma, LUSC lung squamous cell carcinoma, READ rectum adenocarcinoma, OV ovarian serous cystadenocarcinoma, UCEC uterine corpus endometrioid carcinoma. b DNA copy number variation of CREPT was found in 48.28% (56/116) CRC cases by real-time PCR with a Taqman probe targeting CREPT. c The correlation between copy number changes of CREPT and mRNA overexpression (R = 0.485, P < 0.0001)
CREPT promotes CRC cell proliferation and tumorigenesis
To elucidate the function of CREPT in CRC, we examined the effects of CREPT on growth characteristics of colon cancer cells by MTT assay and colony formation assay. CREPT was overexpressed in DLD-1 and HCT116 cell lines that showed low CREPT expression and knockdown in HT29 and SW480 that showed high CREPT expression (S Fig. 1a). The efficiency of overexpression (Fig. 3a) and knockdown (Fig. 3b) of CREPT were verified by RT-PCR and western blot. Ectopic expression of CREPT significantly accelerated cell growth (Fig. 3c) and increased clonogenicity (Fig. 3d) in DLD-1 and HCT116 cells compared with empty vector transfected control cells. In contrast, knockdown of CREPT attenuated cell growth (Fig. 3e) and inhibited clonogenicity (Fig. 3f) in HT29 and SW480 cells. Furthermore, CREPT overexpression significantly accelerated the growth of two normal human colonic epithelial cells HCEC 1CT and 2CT (Fig. 3g). Soft agar assay revealed that overexpression of CREPT in transfected HCT116 cells dramatically enhanced their colony-formation ability (Fig. 3h, S Figure 2). To further assess the function of CREPT on tumorigenesis in vivo, we performed a xenograft model assay through subcutaneously injecting CREPT-overexpressing and control HCT116 cells or SW480 cells with CREPT knockdown and control cells, respectively. As shown in Fig. 3i1, the growth of tumor volume by CREPT-overexpressing cells was significantly increased compared with control cells (P < 0.01). The net weight of tumors formed by CREPT-overexpressing cells was also significantly increased compared with controls at termination of the experiment (P < 0.01; Fig. 3i2). Immunohistochemistry showed that protein expression of CREPT was greatly enhanced in xenograft tumors of the CREPT-overexpressing group compared with control group (Fig. 3i3). The tumor-promoting effect of CREPT in vivo was further confirmed in the knockdown assay of CREPT in nude mice (Fig. 3j).
CREPT promoted colon cancer cell growth and tumor metastasis. a Expression levels of CREPT were increased after transfection with CREPT plasmids in colon cancer cell lines DLD-1 and HCT116 by RT-PCR and western blotting. b Knockdown efficiency of CREPT in HT29 and SW480 cell was confirmed by RT-PCR and western blotting. c Ectopic expression of CREPT significantly enhanced cell viability in both cell lines by MTT assay. d The number of colonies increased when transfected with CREPT plasmids in DLD-1 and HCT116. e Knockdown of CREPT significantly inhibited cell viability in HT29 and SW480 cell lines. f The number of colonies dramatically reduced after knockdown of CREPT in HT29 and SW480 cell lines. g Ectopic expression of CREPT significantly promoted cell viability in HCEC 1CT and 2CT cells. h CREPT overexpression accelerated HCT116 cells colony formation in soft agar. i Ectopic expression of CREPT in HCT116 cells promoted tumor growth and tumor weight in xenograft model. j Knockdown of CREPT in SW480 cells repressed tumor growth and tumor weight. IHC staining showed CREPT expression in xenograft tumor. n = 5. *P < 0.05, **P < 0.01
CREPT enhances invasion and migration abilities of CRC cells
We also investigated the effect of CREPT on metastasis in trans-well migration and invasion assays. Ectopic expression of CREPT significantly increased cell migration (Fig. 4a) and invasion in HCT116 cells (Fig. 4b) compared with control. In contrast, knockdown of CREPT by shRNA dramatically reduced the migratory (Fig. 4c) and invasive abilities (Fig. 4d) of SW480 cells. To further valuate the ability of CREPT on metastasis in vivo, we intravenously injected HCT116 cells with overexpression of CREPT or empty vector in nude mice. After 6 weeks of injection, mice were killed and the lungs were collected. The number of metastasis nodules in lungs was significantly increased in mice injected with CREPT overexpression cells than in mice injected with control cells (P < 0.05) (Fig. 4e). Therefore, CREPT had an important role in colon cancer metastasis.
CREPT enhanced CRC metastasis in vitro and in vivo. Ectopic expression of CREPT-promoted cell invasion a and migration b in HCT116 cells. Knockdown of CREPT suppressed cell invasion c and migration d in SW480 cells. e Ectopic expression of CREPT accelerated lung metastasis of HCT116 cells injected via tail vein. Pictures on the left showed representative lung morphology. HE staining showed representative lungs section. Histograph showed statistic result of metastasis nodules in nude mice lung tissues. n = 5, * P < 0.05, ** P < 0.01
CREPT promotes Wnt/β-catenin signaling pathway in CRC
To probe the pathways regulated by CREPT in CRC, we performed RNA-seq on HCT116 cells stably transfected with CREPT or control vector. KEGG Pathway Enrichment Analysis revealed alterations in several pathways, especially WNT signaling pathway (P < 0.01, Fig. 5a, detailed gene list in S Table 2). Notably, a considerable number of differential genes were well-established WNT/β-catenin downstream targets, including CCND1, c-MYC, and AXIN2. Wnt-response luciferase reporter assay further confirmed this finding (Fig. 5b). Ectopic expression of CREPT significantly enhanced the activity of Wnt signaling reporter Top-Flash by 2.67-fold (Fig. 5b1), while knockdown of CREPT attenuated the activity of Top-flash by 50% (P < 0.01) (Fig. 5b2). We also validated the expression of these three key Wnt signaling downstream targets by RT-PCR and western blot in CREPT overexpressing and knockdown cells. The mRNA and protein expression of CCND1, c-MYC, and AXIN2 were evidently enhanced in CREPT-overexpressing DLD-1, HCT116, HCEC 1CT and 2CT cells compared to the control cells (Fig. 5c, e1, 2); while knockdown CREPT in HT-29 and SW480 cells showed reduced expression (Fig. 5d, e3).
CREPT regulated Wnt/β-catenin signaling pathway in CRC cells. a KEGG analysis based on RNA-sequencing data of HCT116 cells stably overexpressing CREPT or control cells. b Ectopic expression of CREPT in DLD-1 cell line enhanced TopFlash luciferase reporter activity but not FopFlash reporter (b1), while depletion of CREPT reduced dual-luciferase reporter assays in SW480 cells (b2). TopFlash, a classic Wnt-response luciferase reporter containing 3xTCF4 binding site. FopFlash, a negative reporter of which TCF4 binding site is mutant. c The mRNA expression of Wnt downstream targets CCND1, c-MYC, and AXIN2 were enhanced in the presence of CREPT in DLD-1 cells and HCT116 cells. d The mRNA expression of CCND1, c-MYC, and AXIN2 was diminished under CREPT knockdown in HT29 and SW480 cells. e The protein expression of β-catenin, c-MYC, CYCLIN D1, and AXIN2 was increased upon ectopic expression of CREPT in DLD1, HCT116 (e1), HCEC 1CT, and 2CT cells (e2), while decreased in CREPT knockdown HT29 and SW480 cells (e3).
p300 is identified as a novel crucial co-partner of CREPT
Our group previously performed an immunoprecipitation/mass spectrometry experiment in HEK293T cells overexpressing CREPT and identified RNAPII as an interacting protein of CREPT to stimulate CCND1 expression and promote oncogenesis [10]. Using this method, p300 was also identified as a potential CREPT interacting partner (S Fig. 3a). The interaction of CREPT with p300 was further confirmed by transient transfection of Myc-tagged CREPT and HA-tagged p300 followed by co-immunoprecipitation with an anti-HA antibody (Fig. 6a). Furthermore, co-immunoprecipitation using endogenous anti-p300 antibody showed that CREPT had a strong binding to endogenous p300 in SW480 cells (Fig. 6b). To elucidate the role of p300 in CREPT-mediated tumorigenesis, we silenced p300 in CREPT-overexpressed cells (Fig. 6c) and examined the effect on cell growth and colony-formation ability. As shown in Fig. 6d, e, CREPT significantly promoted DLD-1 cell growth, whereas it was significantly dampened after p300 knockdown, suggesting that p300 was an important mediator in CREPT-promoted tumorigenesis. Increasing evidence showed that β-catenin recruits co-activator p300 to form a transcriptionally active complex [22,23,24,25]. The binding of p300 has an important effect on Wnt/β-catenin signaling, leading to differential regulation of target gene expression and different functional outcomes [26, 27]. As a critical partner of CREPT, p300 might be an important co-regulator of CREPT-mediated Wnt/β-catenin signaling activation. To test this hypothesis, we performed a luciferase reporter assay to evaluate the transcriptional activity of β-catenin/TCF4 complex under different transfection conditions. CREPT overexpression significantly increased the activity of Top-flash, whereas co-transfection of CREPT and p300 further enhanced the transcriptional activity of β-catenin/TCF4 complex (Fig. 6f). Furthermore, co-immunoprecipitation assay results showed that β-catenin increased its affinity for TCF4 when cells were transfected with CREPT alone or p300 alone (Fig. 6g, lanes 3 and 4). Furthermore, the interaction between β-catenin and TCF4 was stronger when cells were co-transfected with CREPT and p300 (Fig. 6g, lane 5). Moreover, CREPT-mediated Wnt/β-catenin signaling activation was significantly inhibited after p300 knockdown (Fig. 6h, i), further confirming that CREPT cooperated with p300 in promoting Wnt/β-catenin signaling activation.
p300 was a novel co-partner of CREPT and participated in CREPT-mediated tumorigenesis. a Co-immunoprecipitation showed the interaction between CREPT and p300. Anti-HA antibody was used to immunoprecipitate HA-tagged p300 from whole cell extracts prepared from HEK293T cells co-expressing Myc-CREPT and HA-p300. b Endogenous CREPT was capable of interacting with endogenous p300 in SW480 cells. c p300 knocking down efficiency was examined by real-time PCR and semi-quantitative PCR in DLD1 cells. d DLD1 cell growth rate in p300 knockdown was slower after CREPT overexpression. e Colony-formation assay indicated p300 knockdown cells showed decreased clonogenicity after CREPT overexpression in DLD1 cells. f Co-transfection of CREPT and p300 further enhanced the transcriptional activity of β-catenin/TCF4 complex by dual-luciferase reporter assays. g The β-catenin-TCF4 complex formation was significantly enhanced to the highest degree when cells were co-transfected with CREPT and p300. h The transcriptional activity of β-catenin/TCF4 complex was decreased in p300 knockdown DLD-1 cells with CREPT overexpression. i The mRNA (i1) and protein expression (i2) of WNT/β-catenin signaling downstream targets CCND1, c-MYC, and AXIN2 were decreased in p300 knockdown DLD-1 cells with CREPT overexpression
CREPT enhances the interaction between p300 and β-catenin and promotes p300-mediated acetylation and stability of β-catenin
To further explore the mechanism how CREPT cooperates with p300 to stimulate Wnt signaling, we examined the effect of CREPT on the formation of β-catenin and p300 transcriptional complex by immunoprecipitation assay. As shown in Fig. 7a, a clear increased associated β-catenin was observed in p300 precipitates from CREPT-overexpressing cells, whereas depletion of CREPT attenuated the interaction between p300 and β-catenin (Fig. 7b), indicating that the interaction between p300 and β-catenin was enhanced in the presence of CREPT. p300 is capable of functioning as acetyltransferase for nonhistone targets [28,29,30,31]. p300/CBP coactivator family-mediated acetylation of β-catenin increased its stability and the interaction between β-catenin and TCF4, contributing to β-catenin-mediated neoplastic transformation [22,23,24, 32,33,34]. Thus, we proposed that CREPT might regulate β-catenin acetylation by recruiting p300. Our results showed that β-catenin acetylation was significantly enhanced in CREPT-overexpressing cells (Fig. 7c), but decreased in CREPT knockdown cells (Fig. 7d). Especially, acetylation of endogenous β-catenin was increased in DLD-1 cells with CREPT overexpression (Fig. 7e), whereas decreased in SW480 CREPT knockdown cells (Fig. 7f). To further confirm whether CREPT affects β-catenin stability, we examined the degradation rate of β-catenin after cycloheximide treatment. The results showed that the protein level of β-catenin was significantly higher in the presence of CREPT (Fig. 7g). Therefore, our findings suggested that CREPT-mediated Wnt/β-catenin signaling activation is through cooperating with p300 and promoting β-catenin acetylation and stability.
p300 promoted acetylation and stability of β-catenin and regulated the epigenetic changes on the promoter of Wnt target genes. a CREPT enhanced the interaction between p300 and β-catenin. b Depletion of CREPT diminished the interaction between p300 and β-catenin. c The acetylation level of ectopic β-catenin was greatly enhanced in DLD-1 cells co-transfected with CREPT and p300. d CREPT knockdown in SW480 cells led to reduced acetylation level of ectopic β-catenin induced by p300. e CREPT overexpression caused increased acetylation level of endogenous β-catenin in DLD-1 cells. f The acetylation level of endogenous β-catenin was reduced in CREPT knockdown SW480 cells. g CREPT reduced the rate of β-catenin degradation after cycloheximide addition. h CREPT upregulated the level of H3K27ac and H4ac and downregulated the level of H3K9me3 on the c-MYC promoter region in DLD-1 cells. The relative fold enrichment of H3K27ac, H4ac, and H3K9me3 at the c-MYC promoter region was determined by ChIP analysis using chromatin prepared from CREPT-overexpressing and control DLD-1 cells. i CREPT depletion led to decreased level of H3K27ac and H4ac and elevated level of H3K9me3 in the c-MYC promoter region in SW480 cells. j Proposed mechanistic model of oncogenic function of CREPT in colorectal cancer
CREPT enhances active epigenetic modification of histone in the promoter of Wnt downstream targets
p300 belongs to histone acetyltransferase and regulates transcription via histone acetylation and chromatin remodeling, thus giving an epigenetic tag for transcriptional activation [35, 36]. In this regard, the level of active histone acetylation marker H3K27ac and H4Ac and repressive histone marker H3K9me3 was examined in the promoter of Wnt target gene c-MYC, under CREPT overexpression and knockdown by quantitative ChIP-PCR. Our result showed that CREPT enhanced the level of H3K27ac and H4Ac (Fig. 7h) and reduced level of H3K9me3 in the c-MYC promoter region (Fig. 7i), indicating that p300 recruited by CREPT on Wnt/β-catenin downstream target genes and control the transcription efficiency through directly regulated histone modifications.
CREPT is positively correlated with Wnt signaling pathway in CRC patients
To further prove the clinical relevance of CREPT in CRC patients, we checked the protein expression of CREPT, β-catenin, and C-MYC in our cohort of CRC patients by IHC staining. Pearson correlation analysis revealed that the expression of CREPT significantly correlated with the expression of β-catenin, and C-MYC in CRC clinical samples (P < 0.001, Fig. 8a). Next, we performed Gene Set Enrichment Analysis (GSEA) analysis in CRC patients from TCGA based on RNA-seq data, revealing that CREPT mRNA expression was significantly positively correlated with WNT signaling and three WNT signaling-related pathways of cell cycle, MYC, and VEGF (Fig. 8b). Moreover, we verified mRNA expression levels of key WNT signaling target genes in CRC (refer to Wnt homepage: https://web.stanford.edu/group/nusselab/cgi-bin/wnt/) from TCGA CRC RNA-seq data (Fig. 8c1). We found that CCND1, C-MYC, AXIN2, CLDN1, MET, FGF18, VEGFA, and MSL1 were significantly positively correlated with CREPT mRNA expression (Fig. 8c2). These results indicated that CREPT promotes WNT signaling in CRC patients.
CREPT was positively correlated with Wnt signaling pathway in CRC patients. a Representative IHC pictures to show staining of CREPT, β-catenin and c-MYC using same lot of TMA (a1). Correlation of CREPT with β-catenin or c-MYC was performed according to log10 of H score of individual protein staining (a2). b GSEA results using TCGA colorectal cancer RNA sequencing data indicated CREPT was positive correlated with Wnt signaling. c Correlation of CREPT with Wnt-targeted genes. c1 Heatmap showed expression of CREPT and Wnt-targeted genes in TCGA. c2 Correlation analysis of CREPT with Wnt-targeted genes in TCGA
Discussion
In this study, we found CREPT expression was frequently upregulated in colon cancer cell lines and primary CRC tissues. High level of CREPT was associated with poor disease-free survival for patients with CRC. Copy number gain of CREPT is one of the main regulatory mechanisms of CREPT upregulation in CRC. CREPT was located on chromosome 20q11.23, which is one of the most frequently amplified regions in CRC [16, 17]. Gene amplification is thought to promote overexpression of genes favoring tumor development [6,7,8,9]. CREPT copy number gain or amplification was observed in 48.28% (56/116) CRC cases, indicating that amplification of CREPT is a common event in colorectal tumorigenesis. Importantly, we revealed that copy number gain of CREPT was positively associated with its mRNA overexpression in 116 CRC cases (P < 0.0001), suggesting that increased CREPT copy number contributes to the upregulation of CREPT, which might play an important oncogenic role in CRC.
CRC patients greatly vary in clinical outcome depending on growth status and aggressiveness of the tumors. Currently, TNM stage is the most popularly used as clinical prognostic indicator of disease outcome. However, a part of CRC patients at the early stages of TNM still suffered from disease recurrence. Therefore, additional prognostic biomarkers are requested for better risk assessment. In this regard, we examined the influence of CREPT upregulation on clinical outcomes of 152 CRC patients. High level of CREPT was significantly associated with poor disease-free survival especially at the early stage of patients with CRC (Fig. 1g). Multivariate Cox regression analyses indicated that upregulated CREPT could be regarded as an new independent prognostic marker for CRC recurrence (Table 1).
We previously demonstrated that CREPT was associated with transcriptional complexes not only in promoter regions but also in gene bodies and near mRNA 3′-ends [10]. On one hand, CREPT serves as RNA polymerase II (RNAPII) carboxyl-terminal domain (CTD) scaffolds to coordinate RPAP2-mediated dephosphorylation of phospho-S5, which might be a prerequisite for further association of transcription elongation/processing factors [18, 19]. On the other hand, CREPT was capable of binding to the promoter of CCND1 as well as its 3′-end, thereby promoting loop formation to bring RNAPII physically close to the transcription start site and enhancing RNAPII recycling from the termination to the promoter region [10]. We also reported that the main oncogenic pathway regulated by CREPT is Wnt/β-catenin signaling pathway [20]. However, the structure analysis of CREPT showed that CREPT consists of RPR domain and CCT domain, but lacks common catalyse domain, such as kinase, E3 ligase and acetyltransferase etc. The function of CREPT should thus rely on other functional factor(s). As expected, p300, a novel CREPT interacting protein, was identified as a functional co-player of CREPT in colon cancer by immunoprecipitation and mass spectrometry. Our results showed that besides RNAPII transcriptional complexes, CREPT also coordinated with β-catenin and p300 to control Wnt pathway and its target genes including CCND1, c-MYC, and AXIN2. CREPT and p300 synergized to stimulate Wnt signaling and promote tumor cell proliferation and metastasis in colon cancer cells both in vivo and in vitro. As evidenced by p300 knockdown assay in CREPT-overexpressing cells, CREPT-mediated oncogenic functions and Wnt signaling activation were alleviated by p300 knockdown, suggesting that p300 acted as an essential co-partner of CREPT. More and more evidence demonstrated that p300 and p300/CBP coactivator family members play critical roles in regulating Wnt/β-catenin pathway through directly binding to β-catenin [22,23,24, 32,33,34]. As acetyltransferases, p300 and p300/CBP coactivator family members catalyze the acetylation of β-catenin, thereby improving β-catenin stability, increasing the affinity of β-catenin for TCF4 and leading to efficient transcription [24, 33, 34]. The acetylation of Lys345 on β-catenin by p300 increased the affinity of β-catenin for TCF4 [24]. The cooperation between p300 and β-catenin was greatly dampened by the K345R mutation, implying that acetylation of β-catenin plays an important role in efficient activation of WNT/β-catenin transcription. Interestingly, acetylation of Lys345 on β-catenin was strongly enriched in the colon cancer cells with hyperactivating Wnt/β-catenin signaling [24]. p300 was required for β-catenin-mediated neoplastic transformation in colon cancer [23]. In our study, immunoprecipitation result showed that CREPT promoted the association of β-catenin with p300 and enhanced β-catenin acetylation. The transcriptional activity of β-catenin/TCF4 complex and the binding affinity of β-catenin for TCF4 were increased to the higher degree after co-transfection of CREPT and p300. Moreover, the degradation rate of β-catenin was slowed down in the presence of CREPT after cycloheximide treatment, implying that CREPT affected the level of β-catenin via a post-translational mode due to enhanced β-catenin acetylation. Therefore, one of the key mechanisms by CREPT is that it coordinates with p300 to enhance the interaction between p300 and β-catenin, causing acetylation of β-catenin and robust activation of Wnt/β-catenin pathway. Moreover, p300 is also a histone acetyltransferase. It acted not only on β-catenin but also on chromatin directly. We found that CREPT modulated the level of active histone acetylation marker H3K27ac and H4Ac and repressive histone marker H3K9me3 in the promoter of Wnt target genes. Collectively, this study revealed the mechanisms underlying CREPT in regulating Wnt signaling pathway: CREPT recruited p300 to enhance the acetylation and stability of β-catenin and thus activating Wnt signaling pathway.
In summary, we have identified a novel functional oncogene CREPT overexpressed in CRC. Upregulated CREPT was associated with poor disease-free outcome. Copy number gain of CREPT contributes to its upregulation in CRC. CREPT promoted CRC growth and invasion/migration ability through coordinating with a novel interacting partner p300. CREPT and p300 synergized to stimulate Wnt/β-catenin signaling and promoted tumorigenesis in two ways (Fig. 7j). On one hand, CREPT enhanced the interaction between p300 and β-catenin and promoted p300-mediated acetylation of β-catenin, leading to increased β-catenin-TCF4 complex formation on Wnt target genes. On the other hand, p300 recruited by CREPT directly regulated the histone modifications on the promoter of Wnt target genes. Collectively, CREPT plays a pivotal oncogenic role in colorectal carcinogenesis through promoting Wnt/β-catenin pathway via cooperating with p300. CREPT may serve as a prognosis biomarker for CRC patients.
Materials and methods
Plasmids and reagents
Myc-CREPT, Flag-β-catenin, HA-p300, HA-TCF4, Top-Flash, Fop-Flash constructs were used in this study. Anti-cyclinD1 (A-12), anti-Myc (9E10), anti-HA (F-7), anti-β-catenin (E-5), anti-Ac-lysine (7F8), and anti-GFP (FL) antibodies and protein G-agarose beads (sc-2002) were from Santa Cruz Biotechnology, Inc (Dallas, Texas, USA). Anti-c-Myc (9402), anti-AXIN2 (2151) used for western blot was purchased from Cell Signaling Technology, Inc (Danvers, MA). Anti-Flag (F1804), anti-β-Actin (AC-15), Crystal Violet (C3886), DMSO (D2650), and MTT (M5655) were purchased from Sigma-Aldrich (St Louis, MO). Anti-c-MYC (ab32072) for IHC, anti-H3K27ac (ab4729), anti-H3K9me3 (ab8898) antibodies were purchased from Abcam (Cambridge, MA, USA). Anti-H4ac (06-866) antibody was from Millipore (Hong Kong, China). Anti-β-catenin (M33539) for IHC was from Dako/Agilent Technologies (USA). Anti-CREPT antibody was generated by our lab. siRNAs were synthesized by Shanghai GenePharma Co. Ltd, China. The oligo sequence information as: si-p300 #1: GGACUACCCUAUCAAGUAATT; si-p300 #2: CAUCACGGGUAUACAAAUATT; si-CREPT: GCAAGAACGAAGUGUUAUTT; siRNA negative control (NC): UCUCCGAAGUCACGUTT. siRNAs were transfected into cells using Lipofectamine™ RNAiMAX Transfection Reagent (13778150) twice. FastStart Universal SYBR Green Master used for real-time PCR was purchased from Roche Applied Sciences (Indianapolis, IN, USA). Puromycin (A1113803) and Hygromycin B (10687010) was from ThermoFisher Scientific (Waltham, MA). Cycloheximide (CHX) was from AMRESCO Inc. (Ameresco, Ohio).
Cell lines
Human CRC cell lines (CaCO2, Colo205, DLD-1, HCT116, HT29, LoVo, SW1116, SW480, and SW620) and one immortalized colorectal epithelial cell line (NCM460) were from the American Type Culture Collection (Manassas, VA) and were cultured as described in previous study [37]. HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS. Cells were kept at 37 °C in a 5% CO2-containing atmosphere. Human colonic epithelial cells (HCEC) 1CT and 2CT were a gift from Prof. Jerry W. Shay (University of Texas Southwestern Medical Center, Dallas, Texas, USA) and were cultured in the conditions previously reported [21]. For generating CREPT stable overexpression or knockdown cell lines, cells were infected with lentivirus and then selected by puromycin at different concentration: 2 μg/ml (DLD-1), 1.5 μg/ml (HCT116), 2 μg/ml (HT29), 4 μg/ml (SW480) or by Hygromycin B (400 μg/ml) for HCEC 1CT and 2CT.
Clinical samples
Thirteen primary paired CRC samples (tumor sample and adjacent non-tumorous sample) randomly selected from Prince of Wales Hospital (Hong Kong) were used for western blot detection. A tissue microarray including 152 cases of CRC patient samples from Prince of Wales Hospital was used for immunohistochemistry staining. In total, 116 CRC tissues obtained from the Beijing University Cancer Hospital (China) were used for copy number variant assay [38]. All patients signed informed consent before specimen collection. The Joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee and Human Ethics Committee of Beijing University Cancer Hospital approved the ethics individually.
Animal assay
For xenograft model, 5×106 cells were subcutaneously injected into the right or left dorsal flank of each 4-week-old male Balb/c nude mice, respectively. Each experimental group consisted of five mice. Tumor volume was measured every 2 days for 4 weeks and calculated using the formula V = (W2 × L)/2 (V = volume, L = length, W = width). For the metastasis model, 2×106 cells were intravenously injected through the tail vein of nude mouse (4-week old, 5 mice/group). All mice were killed at 6 weeks after injection. The lungs from each mouse were embedded in paraffin for hematoxylin and eosin staining. All experimental procedures were approved by the Animal Ethics Committee of the Chinese University of Hong Kong.
Copy number assay
Gene copy number variation was performed using a TaqMan copy number assay (ThermoFisher Scientific) according to the manufacturer’s instructions. Briefly, genomic DNA was extracted from CRC samples as templates of PCR. DNA copy number was examined by real-time PCR using a specific Taqman probe against CREPT (Hs01601842_cn) and a reference probe targeted to RNase P (#4403326). Each sample was detected with four replicates to generate the reliable copy number calls. The result was analyzed using Applied Biosystems® CopyCaller® Software (http://www.appliedbiosystems.com/absite/us/en/home/support/software/real-time-pcr/copycaller.html).
Cell proliferation analysis
MTT assay was used for cell proliferation detection. 2000 cells/well were seeded into a 96-well plate. The absorbance at 570 nm was measured using Multiskan™ GO Microplate Spectrophotometer (ThermoFisher Scientific). Three independent experiments were performed.
Colony formation
For anchorage-dependent growth, 1000 cells were seeded into a six-well plate. For anchorage-independent growth in soft agar, 7000 cells were inoculated in cell culture medium containing 0.35% agar and seeded in six-well plate with medium containing 0.7% agar. Cells were grown for 2 weeks (anchorage-dependent) or 4 weeks (anchorage-independent). Cell clones were counted and presented as mean ± standard deviation (SD) from three individual experiments.
Cell migration and invasion assay
BD Falcon Cell Culture Inserts (#353097) purchased from BD (Franklin Lakes, New Jersey, USA) were used for migration assay. For invasion assay, BD BioCoat™ Matrigel™ Invasion Chamber (BD, #354480) was used according to the manufacturer’s instructions. The number of invaded cells was counted under a phase contrast microscope. Cells in five different fields of each well were averaged.
RNA sequencing
HCT116 cells overexpressing CREPT or empty vector were harvested for total RNA extraction. RNA sequencing and analysis were performed by Beijing Novogene Technology Co. Ltd.
Protein half-lives assay
To detect the half-life of the β-catenin protein, Flag-β-catenin was transfected into HEK293T cells alone or with Myc-CREPT. After 24 h of transfection, cells were treated with 50 μg/ml cyclohexanone (CHX) and further cultured for indicated time course. Cells lysates were examined using western blotting.
Co-immunoprecipitation and western blotting
Co-immunoprecipitation was performed as previously reported [39]. Specifically, for transient transfection interaction assays, Flag-β-catenin, HA-TCF4, HA-p300 and Myc-CREPT plasmids were transfected into HEK293T cells according to the assay design. For endogenous interaction assays, the nuclear fraction of SW480 cells was used for immunoprecipitation
Luciferase assay
HEK293T, DLD-1, or SW480 cells were transiently transfected with the indicated plasmids using Lipofectamine 2000. Luciferase activity was measured after 24 h of transfection using a Dual-Luciferase reporter assay system (E1910, Promega, Madison, WI). The luciferase activity was normalized by firefly against Renilla luciferase activity and presented as mean ± SD.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as previously described [20]. Briefly, sonicated DNA was immunoprecipitated with histone modification markers H3K27Ac, H4Ac, and H3K9me3 antibodies. Normal IgG was used as negative control. ChIPed DNA was examined by real-time PCR to evaluate histone modification status on the c-MYC promoter, using primers of TCF4 binding site: 5′-TTGCTGGGTTATTTTAATCAT-3′ and 5′-ACTGTTTGACAAACCGCATCC-3′ [40]. The ChIP-qPCR data was normalized using Percent Input Method.
Statistical analysis
All experiments were repeated at least three times. Data were presented as mean ± standard deviation. Correlation analysis of copy number and mRNA was performed using linear regression. Significant differences between groups were determined using the Student’s ttest. The difference between the two groups in cell growth curve and tumor growth rate of nude mice were determined by repeated-measures analysis of variance. The H-score of CREPT as a weighted sum of the intensity of IHC tumor cell nuclei was calculated as reported [41]. Survival analysis was calculated using Kaplan–Meier analysis.
References
Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, et al. Colorectal cancer. Lancet. 2010;375:1030–47.
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108.
Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13.
Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507.
Xie T, G DA, Lamb JR, Martin E, Wang K, Tejpar S, et al. A comprehensive characterization of genome-wide copy number aberrations in colorectal cancer reveals novel oncogenes and patterns of alterations. PLoS ONE. 2012;7:e42001.
Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, et al. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996;15:234–45.
Meijer GA, Hermsen MA, Baak JP, van Diest PJ, Meuwissen SG, Belien JA, et al. Progression from colorectal adenoma to carcinoma is associated with non-random chromosomal gains as detected by comparative genomic hybridisation. J Clin Pathol. 1998;51:901–9.
Douglas EJ, Fiegler H, Rowan A, Halford S, Bicknell DC, Bodmer W, et al. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004;64:4817–25.
Nakao K, Mehta KR, Fridlyand J, Moore DH, Jain AN, Lafuente A, et al. High-resolution analysis of DNA copy number alterations in colorectal cancer by array-based comparative genomic hybridization. Carcinogenesis. 2004;25:1345–57.
Lu D, Wu Y, Wang Y, Ren F, Wang D, Su F, et al. CREPT accelerates tumorigenesis by regulating the transcription of cell-cycle-related genes. Cancer Cell. 2012;21:92–104.
Jung HM, Choi SJ, Kim JK. Expression profiles of SV40-immortalization-associated genes upregulated in various human cancers. J Cell Biochem. 2009;106:703–13.
She Y, Liang J, Chen L, Qiu Y, Liu N, Zhao X, et al. CREPT expression correlates with poor prognosis in patients with retroperitoneal leiomyosarcoma. Int J Clin Exp Pathol. 2014;7:6596–605.
Wang Y, Qiu H, Hu W, Li S, Yu J. RPRD1B promotes tumor growth by accelerating the cell cycle in endometrial cancer. Oncol Rep. 2014;31:1389–95.
Ma J, Ren Y, Zhang L, Kong X, Wang T, Shi Y, et al. Knocking-down of CREPT prohibits the progression of oral squamous cell carcinoma and suppresses cyclin D1 and c-Myc expression. PLoS ONE. 2017;12:e0174309.
Zheng G, Li W, Zuo B, Guo Z, Xi W, Wei M, et al. High expression of CREPT promotes tumor growth and is correlated with poor prognosis in colorectal cancer. Biochem Biophys Res Commun. 2016;480:436–42.
Carvalho B, Postma C, Mongera S, Hopmans E, Diskin S, van de Wiel MA, et al. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89.
Xu L, Li X, Cai M, Chen J, Li X, Wu WK, et al. Increased expression of Solute carrier family 12 member 5 via gene amplification contributes to tumour progression and metastasis and associates with poor survival in colorectal cancer. Gut. 2016;65:635–46.
Ni Z, Olsen JB, Guo X, Zhong G, Ruan ED, Marcon E, et al. Control of the RNA polymerase II phosphorylation state in promoter regions by CTD interaction domain-containing proteins RPRD1A and RPRD1B. Transcription. 2011;2:237–42.
Ni Z, Xu C, Guo X, Hunter GO, Kuznetsova OV, Tempel W, et al. RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. Nat Struct Mol Biol. 2014;21:686–95.
Ren F, Wang R, Zhang Y, Liu C, Wang Y, Hu J, et al. Characterization of a monoclonal antibody against CREPT, a novel protein highly expressed in tumors. Monoclon Antib Immunodiagn Immunother. 2014;33:401–8.
Roig AI, Eskiocak U, Hight SK, Kim SB, Delgado O, Souza RF, et al. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology. 2010;138:1012–21 e1011−1015.
Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 2000;19:1839–50.
Sun YN, Kolligs FT, Hottiger MO, Mosavin R, Fearon ER, Nabel GJ. Regulation of beta-catenin transformation by the p300 transcriptional coactivator. Proc Natl Acad Sci USA. 2000;97:12613–8.
Levy L, Wei Y, Labalette C, Wu YF, Renard CA, Buendia MA, et al. Acetylation of beta-catenin by p300 regulates beta-catenin-Tcf4 interaction. Mol Cell Biol. 2004;24:3404–14.
Ma H, Nguyen C, Lee KS, Kahn M. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene. 2005;24:3619–31.
Mosimann C, Hausmann G, Basler K. beta-Catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10:276–U257.
Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. EMBO J. 2012;31:2714–36.
Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606.
Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20:1331–40.
Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–73.
Inuzuka H, Gao D, Finley LW, Yang W, Wan L, Fukushima H, et al. Acetylation-dependent regulation of Skp2 function. Cell. 2012;150:179–93.
Takemaru KI, Moon RT. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol. 2000;149:249–54.
Wolf D, Rodova M, Miska EA, Calvet JP, Kouzarides T. Acetylation of beta-catenin by CREB-binding protein (CBP). J Biol Chem. 2002;277:25562–7.
Ge XJ, Jin QH, Zhang F, Yan TT, Zhai QW. PCAF acetylates beta-catenin and improves its stability. Mol Biol Cell. 2009;20:419–27.
Dancy BM, Cole PA. Protein lysine acetylation by p300/CBP. Chem Rev. 2015;115:2419–52.
Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–73.
Wang S, Dong Y, Zhang Y, Wang X, Xu L, Yang S, et al. DACT2 is a functional tumor suppressor through inhibiting Wnt/beta-catenin pathway and associated with poor survival in colon cancer. Oncogene. 2015;34:2575–85.
Zhang J, Tsoi H, Li X, Wang H, Gao J, Wang K, et al. Carbonic anhydrase IV inhibits colon cancer development by inhibiting the Wnt signalling pathway through targeting the WTAP-WT1-TBL1 axis. Gut. 2015;65:1482–93.
Zhang Y, Liu C, Duan X, Ren F, Li S, Jin Z, et al. CREPT/RPRD1B, a recently identified novel protein highly expressed in tumors, enhances the beta-catenin.TCF4 transcriptional activity in response to Wnt signaling. J Biol Chem. 2014;289:22589–99.
Ito K, Lim AC, Salto-Tellez M, Motoda L, Osato M, Chuang LS, et al. RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell. 2008;14:226–37.
Sun W, Zhang Y, Wong KC, Liu K, Yang Y, Wu B, et al. Increased expression of GATA Zinc Finger Domain Containing 1 via gene amplification promotes liver cancer by directly inducing PRL3. Hepatology 2017. https://doi.org/10.1002/hep.29750.
Acknowledgements
This project was supported by the National Natural Science Foundation (81301775, 81230044, 81572728), RGC-GRF Hong Kong (14106415, 14111216, 14163817), 135 program project (2016YFC1303200), National Key Research and Development Program (2016YFA0500301); Shenzhen Virtual University Park Support Scheme to CUHK Shenzhen Research Institute.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, Y., Wang, S., Kang, W. et al. CREPT facilitates colorectal cancer growth through inducing Wnt/β-catenin pathway by enhancing p300-mediated β-catenin acetylation. Oncogene 37, 3485–3500 (2018). https://doi.org/10.1038/s41388-018-0161-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0161-z
This article is cited by
-
Beyond canonical PROTAC: biological targeted protein degradation (bioTPD)
Biomaterials Research (2023)
-
Roles of non-coding RNAs in the metabolism and pathogenesis of bladder cancer
Human Cell (2023)
-
The interaction of canonical Wnt/β-catenin signaling with protein lysine acetylation
Cellular & Molecular Biology Letters (2022)
-
Differential enrichment of H3K9me3 in intrahepatic cholangiocarcinoma
BMC Medical Genomics (2022)
-
Long non-coding RNA NEAT1 mediated RPRD1B stability facilitates fatty acid metabolism and lymph node metastasis via c-Jun/c-Fos/SREBP1 axis in gastric cancer
Journal of Experimental & Clinical Cancer Research (2022)