Abstract
Glioblastoma multiforme is a brain malignancy characterized by high heterogeneity, invasiveness, and resistance to current therapies, attributes related to the occurrence of glioma stem cells (GSCs). Transforming growth factor β (TGFβ) promotes self-renewal and bone morphogenetic protein (BMP) induces differentiation of GSCs. BMP7 induces the transcription factor Snail to promote astrocytic differentiation in GSCs and suppress tumor growth in vivo. We demonstrate that Snail represses stemness in GSCs. Snail interacts with SMAD signaling mediators, generates a positive feedback loop of BMP signaling and transcriptionally represses the TGFB1 gene, decreasing TGFβ1 signaling activity. Exogenous TGFβ1 counteracts Snail function in vitro, and in vivo promotes proliferation and re-expression of Nestin, confirming the importance of TGFB1 gene repression by Snail. In conclusion, novel insight highlights mechanisms whereby Snail differentially regulates the activity of the opposing BMP and TGFβ pathways, thus promoting an astrocytic fate switch and repressing stemness in GSCs.
Similar content being viewed by others
Introduction
Glioblastoma multiforme (GBM) is a fatal adult brain tumor with median survival of 12–15 months [1]. GBM exhibits heterogeneous differentiation, high invasiveness, vascular density, and resistance to radiotherapy and chemotherapy [2]. GBM cancer stem cells are non-responsive to current treatments and contribute to acquired chemoresistance [3]. These glioma stem cells (GSCs) self-renew and are responsible for tumor initiation, propagation, and relapse after therapy [4]. GSCs resemble neural stem cells, as their fate is controlled by environmental cues such as Notch, Wnt, and the transforming growth factor β (TGFβ) family [5, 6].
The 33 members of the human TGFβ family contain the TGFβ and the bone morphogenetic protein (BMP) subfamilies [7]. These ligands bind to type II receptors (TGFBR2, ACVR2A, ACVR2B, BMPR2, and AMHR2), facilitating recruitment and phosphorylation of the type I receptors, also known as activin receptor-like kinases (ALK: ALK4/5/7 for the TGFβ and ALK1/2/3/6 for the BMP subfamily). Hetero-tetrameric receptors signal via non-SMAD and SMAD pathways. Type I TGFβ receptors phosphorylate receptor-regulated (R-)SMADs, i.e., SMAD2/3, and BMP receptors phosphorylate SMAD1/5/8; activated R-SMADs form complexes with SMAD4 and regulate transcription of genes [7]. In addition, inhibitory (I-) SMADs (SMAD6/7) inhibit these pathways.
Mimicking their functions during normal brain development, TGFβ members play opposing roles in the maintenance of GSCs in human GBM. TGFβ promotes GSC proliferation [8] and self-renewal by inducing leukemia inhibitory factor (LIF) [9] and Sox2 [10] expression. BMPs direct GSC differentiation toward the astrocytic lineage and deplete the GSC pool, acting as tumor suppressors [11,12,13,14]. GSCs are protected from paracrine BMP actions by expressing the extracellular antagonist Gremlin [15]. Clinically, anti-TGFβ therapy in GBM offers promising results [16].
We previously described that BMPs exert their differentiation effects by inducing the transcription factor Snail [13]. Snail promotes epithelial-to-mesenchymal transition (EMT) in embryos or epithelial-derived tumors [17]. In GBM, Snail promotes invasion [13, 18, 19], but mechanisms by which Snail impairs GSC self-renewal and promotes astrocytic fate remain unknown. We have addressed this important question and demonstrate an intriguing feed-forward loop whereby Snail controls the activities of both TGFβ and BMP pathways at the level of secreted ligands and intracellular SMAD function.
Results
Snail promotes astrocytic fate and enhances the BMP pathway
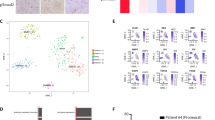
By screening a panel of patient-derived, established (U2987MG, U2990MG, U343MG, M343MGa-Cl2.6) and stem cell lines (U3013MG, U3024MG, U3028MG, U3034MG), we found that U2987MG, U3013MG, U3024MG, U3028MG, and U3034MG cultures contained GSCs, whereas the other GBM cell models did not [13]. We employed U2987MG cells, which exhibited more robust and sustained responses to TGFβ1 and BMP2, BMP4, and BMP7 signaling, compared to the other models (Supplementary Figure S1, where both TGFβ1 and BMP7 induce R-Smad phosphorylation up to 72 h after stimulation and for other gene responses [13]). Transcriptomic analysis in U2987MG cells stably expressing Snail (U2987MG-Snail cells; clones F and G) versus control cells (clones A and B) in duplicate identified 1100 differentially expressed genes. Among the top upregulated genes in the U2987MG-Snail cells were astrocytic differentiation markers, the intermediate filament glial acidic fibrilary protein (GFAP) and the extracellular matrix glycoprotein secreted protein, acidic, cysteine rich-like-1 (SPARCL1) (Fig. 1a, b). In order to corroborate that Snail can induce astrocytic fate in GBMs, we compared the differentially expressed genes in our dataset to those differentially expressed genes in normal mouse astrocytes versus other brain cell lineages (oligodendrocyte progenitors, oligodendrocytes, neurons, cultured astroglia) reported by Cahoy et al. [20] We chose a murine dataset as this one contained several brain cell types and allowed us to discriminate genes expressed in astrocytes from those in other brain cell types. The resulting 130 common genes were plotted in a heatmap relative to the biological samples, named according to cell clone and their respective duplicates (bis, Fig. 1a). GBM-Snail clones expressed very high (GFAP), intermediately high (SPARCL1), and moderately high (GLUL, glutamate-ammonia ligase, another astrocytic marker) levels (Fig. 1b); note the differences in the basal level of expression of these genes (Supplementary Figure S2A).
Snail promotes astrocytic fate switch and impairs self-renewal capacity in GSCs. a Subgroup of genome-wide mRNA expression in U2987MG-Snail (clones F and G) or U2987MG-mock cells (pcDNA3 clones A and B). A heatmap displays differentially expressed genes, common between astrocytes (Cahoy et al. [20]) and U2987MG-Snail clones. Upregulated (red) and downregulated (blue) genes are listed; relevant genes are highlighted. b, c qRT-PCR analysis of mRNA expression (n = 3–6, technical triplicate) presented as mean ± SD. Statistical comparison (one-way Anova) indicates significant differences at: *p < 0.05, **p < 0.01, ***p < 0.001. d Nestin immunoblot with loading control (Gapdh) in cells cultured in complete (FBS) media for 48 h. e ELDA of U2987MG cells (A-pcDNA3, black symbols and F-Snail, red symbols) cultured for 10 days (n = 2, technical octaplicates). The number of wells containing spheres for each plating density is plotted. Steeper slopes indicate higher frequencies of sphere-forming cells. A table indicates average stem cell frequency per cell group
Next, we confirmed that Snail-induced astrocytic differentiation coincided with loss of stem cell and multidrug resistance gene expression, as these were previously linked to GSC stemness and chemoresistance (Fig. 1c, d; Supplementary Figure S2B). Moreover, U2987MG-Snail cells exhibited loss of self-renewal capacity measured by limiting dilution assay of gliomasphere formation (Fig. 1e).
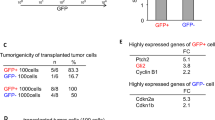
Unexpectedly, among the co-expressed genes in U2987MG-Snail and normal astrocytic cells (Fig. 1a), were inhibitor of differentiation-4 (ID4) and N-myc downstream regulated-1 (NDRG1), both established targets of BMP signaling [21, 22]. Validation of expression of all four ID family members indicated that Snail induced only ID4 (Fig. 2a, Supplementary Figure S3A). Consequently, GSCs expressing Snail exhibited enhanced phospho-SMAD1/5 levels, without changing their total SMAD1/5 (Fig. 2b). Systematic analysis of BMP pathway genes showed that GSCs expressing Snail had diminished expression of three BMP receptors (ALK2, ACVR2B, and BMPRII), whereas ALK6 and ACVR2A did not change significantly (Supplementary Figure S3B, C). Furthermore, BMP receptor expression was not strongly affected upon stable knockdown of endogenous Snail in parental U2987MG cells (Supplementary Figure S3D, E). Interestingly, Snail-expressing cells exhibited lower levels of extracellular (chordin (CHRD), chordin-like-2 (CHRDL2)) and intracellular (SMAD6, SMAD7) negative regulators of BMP signaling (Fig. 2c, e); on the other hand, these cells showed high BMP4/5 mRNA levels and significant upregulation of secreted BMP4 protein in the tumor cell media (Fig. 2d–f). In contrast, BMP2/7 mRNA levels did not significantly change (Supplementary Figure S3F). The transcription factor GATA6 can repress BMP4 expression in colon cancer cells [23]; accordingly, GBM cells expressing Snail showed significantly repressed GATA6 mRNA (Supplementary Figure S3G), which can explain the dramatic derepression of BMP4 observed in these cells (Fig. 2d–f).
Snail expression in GBMs induces BMP signaling, promoting the astrocytic fate switch. U2987MG cells expressing pcDNA3-Snail (clones F and G) or empty vector (clones A and B). a, c, d qRT-PCR analysis of mRNA expression (n = 3, technical triplicate). b Immunoblot analysis for the indicated proteins and loading control Gapdh. Representative experiment out of three. e TGFβ family-related gene expression analyzed by Taqman array is graphed as fold-change of expression in F-Snail versus A-pcDNA cells. A horizontal dotted line indicates the onefold baseline (no change). f Human BMP4 protein levels in conditioned media determined by ELISA (n = 2, technical duplicate). g, h Cells were transiently transfected with siControl (−), siSMAD1 and/or siSMAD5 (+) siRNAs; g qRT-PCR analysis of mRNA expression (n = 2, technical triplicate), h quantification of nuclear pSmad1 staining. i, j Cells treated with or without Noggin (0.25 µg/ml) for 48 h; i qRT-PCR analysis of mRNA expression (n = 2, technical triplicate), j quantification of nuclear pSmad1 staining. Results are expressed as mean ± SD. Statistical comparison (one-way Anova for a, c, d, e; two-way Anova for g–j) indicates significant differences at: *p < 0.05, **p < 0.01, ***p < 0.001
The Snail-induced BMP pathway governs astrocytic fate
In order to elucidate the role of BMP signaling in Snail-induced astrocytic fate determination, we blocked the BMP pathway at multiple levels. Interfering with BMPR activity or expression, diminished GFAP, SPARCL1, and ID4 mRNA and GFAP protein levels, and decreased phospho-SMAD1/5 levels in GSCs expressing Snail (Supplementary Figure S4). Silencing the BMP effectors SMAD1/5 individually or in combination (Supplementary Figure S5A) diminished GFAP and ID4 expression (Fig. 2g, Supplementary Figure S5C) and decreased the nuclear accumulation of phosphorylated-SMAD1/5 (Fig. 2h). However, these specific small interfering RNAs (siRNAs) targeting SMAD1 and SMAD5 had no effect on GSC stem cell gene repression by Snail (Supplementary Figure S5B). To investigate the role of autocrine-secreted BMPs, we treated the cells with the physiological extracellular BMP antagonist Noggin; GFAP mRNA and protein levels and ID4 mRNA expression and nuclear accumulation of phospho-SMAD1/5 levels were diminished (Fig. 2i, j, Supplementary Figure 5D). In agreement with the SMAD1/5 silencing results, Noggin failed to revert the stem cell gene suppression by Snail (Supplementary Figure S5E).
Snail suppresses TGFβ1 signaling. TGFβ family analysis in U2987MG cells expressing pcDNA3-Snail (clones F and G) or the empty vector (clones A and B). a TGFβ family-related gene expression analyzed by Taqman array is graphed as fold-change of expression in F-Snail versus A-pcDNA cells. A horizontal dotted line indicates the onefold baseline (no change). b qRT-PCR analysis of mRNA expression (n = 3, technical triplicate). c Human TGFβ1 protein levels in conditioned media determined by ELISA (n = 2, technical duplicate). d Protein expression analysis of secreted EVs by multiplex PEA graphed as fold-change in F-Snail versus A-pcDNA cells. A horizontal dotted line indicates onefold baseline (no change). e Immunoblot analysis of the indicated proteins and Gapdh (representative experiment). f Immunofluorescence staining of SMAD3 (left) and quantification of nuclear SMAD3 staining (right). Results expressed as mean ± SD. Statistical comparison (one-way Anova) indicates significant differences at: *p < 0.05, ***p < 0.001
Snail suppresses TGFβ1 signaling
TGFβ promotes stem cell properties in glioma [24]. In order to explore whether TGFβ pathways, in addition to BMPs, are affected in GBM cells expressing Snail, we checked the expression levels of TGFβ isoforms and downstream effectors in our microarray (Supplementary Figure S6A); this gave a first indication that TGFβ1 ligand and SMAD3 levels might be downregulated by Snail overexpression (the adjusted p-values though were not significant). We proceeded and analyzed several members of the TGFβ/activin subfamily by a Taqman array (Fig. 3a); TGFB1, but not TGFB2 or TGFB3 expression, was strongly reduced in GBMs expressing Snail. In addition, TGFβ receptors TGFBR2 and TGFBR3 were strongly reduced, whereas TGFBR1 was upregulated; INHA, encoding one of the activin subunits, and the activin extracellular antagonist FST were dramatically upregulated as well as the growth and differentiation factors GDF9 and GDF15 (Fig. 3a). Furthermore, the Taqman array included 19 probes for genes not related to the TGFβ family, whose expression did not exhibit significant changes (Supplementary Figure S6B). TGFB1-3 quantitative reverse transcriptase PCR corroborated the array results; SERPINE1/PAI1, an early response gene of TGFβ1 signaling, was strongly underexpressed, reflecting the downregulation of TGFB1 (Fig. 3b). In order to determine the protein level changes, we interrogated the secretome of U2987MG-Snail cells by analyzing secreted TGFβ1 levels (Fig. 3c) and protein content of extracellular vesicles (EVs) by multiplex proximity extension assay (PEA) (Fig. 3d). GBMs expressing Snail secreted lower SERPINE1 and LAP-TGFβ1 levels, and their conditioned media contained less mature TGFβ1. Concomitantly, total SMAD3, but not SMAD2 protein levels, were decreased as shown by immunoblot and quantitative immunofluorescence analysis (Fig. 3e, f, Supplementary Figure S6C).
Snail forms complexes with SMADs. a Snail ChIP on the TGFB1 promoter in U2987MG cells expressing pcDNA3-Snail (clone F) or the empty vector (clone A) (n = 3, technical triplicate). Results expressed as mean ± SD. Statistical comparison (Student’s t-test) indicates significant differences at: *p<0.05. Rabbit IgG control immunoprecipitation is shown. b Immunoprecipitation of endogenous SMAD2/3 followed by immunoblotting for Snail and SMAD2/3 in U2987MG cells stimulated with vehicle (–) or with 5 ng/ml TGFβ1 for 2 h (+). Negative control immunoprecipitation using non-specific IgG and endogenous total cell lysate (TCL) protein levels before immunoprecipitation are shown. c Pulldown assay of 293T cell extracts expressing pcDNA3-HA-Snail, loaded on purified GST or deletion mutants of GST-SMAD3, followed by GST and HA immunoblotting. Stars show specific SMAD3 constructs and input shows HA-Snail levels prior to pulldown. d Pulldown assay of U2987MG cell extracts expressing pcDNA3-Snail (clone F) or empty vector (clone A), loaded on purified GST or deletion mutants of GST-SMAD1, followed by GST and Snail immunoblotting. Arrows show each SMAD1 construct. TCL shows endogenous protein levels before pulldown. e U2987MG cells expressing pcDNA3-Snail (clone F) or empty vector (clone A) stimulated with vehicle (–) or 30 ng/ml BMP7 for 2 h (+); (left) Snail immunoprecipitation followed by Snail, SMAD1, and SMAD4 immunoblotting, (right) SMAD1 immunoprecipitation followed by Snail, SMAD1, and SMAD4 immunoblotting. TCL shows endogenous protein levels before immunoprecipitation. f U2987MG cells stimulated with vehicle (−BMP7) or 30 ng/ml BMP7 for 2 h. Immunoprecipitation of Snail (left), SMAD1 (middle), and SMAD4 (right) followed by Snail, SMAD1, and SMAD4 immunoblotting. Negative control immunoprecipitation using non-specific IgG is shown. TCL shows the levels of endogenous proteins before immunoprecipitation
Using MAPPER [25], we investigated whether Snail could bind to the TGFB1 promoter. MAPPER predicted a Snail-binding site in position −486 relative to the transcription start site of the TGFB1 gene, next to a SMAD3-binding site. Indeed, Snail did bind to the TGFB1 promoter, as demonstrated by chromatin immunoprecipitation (ChIP; Fig. 4a). We have previously published that Snail can interact with SMAD3/4 during EMT [26]. In GSCs, endogenous SMAD2/3 interacted with Snail in response to TGFβ1 (Fig. 4b). Snail interacted with SMAD2 and SMAD3 also in transfected human embryonic kidney cells (Supplementary Figure S7A). Both N- and C-terminal Snail domains associated with SMAD2/3 (Supplementary Figure S7B, C). The N-terminal Mad-homology (MH) 1 domain of SMAD contains the nuclear localization signal and binds to DNA and many transcription factors; a middle Linker region binds to ubiquitin ligases and regulates SMAD stability; the C-terminal MH2 domain transactivates gene transcription, binds to type I receptors, and the very C-terminal serine residues of R-SMADs are phosphorylated. Snail interacted with deletion mutants lacking either SMAD3 MH1 or MH2 domains (Fig. 4c), suggesting more than one SMAD3 epitopes responsible for binding to Snail. Snail also interacted with SMAD1/5 of the BMP pathway. SMAD1 interacted through its MH1 and Linker domains (Fig. 4d), whereas both N- and C-terminal Snail domains associate with SMAD1/5 (Supplementary Figure S8). Endogenous Snail interacted with endogenous SMAD1 and SMAD4 in U2987MG and in Snail-expressing cells; BMP7 stimulation enhanced complex formation (Fig. 4e, f). Interestingly, silencing of Snail expression selectively impairs TGFβ1 and BMP7 downstream effects. On one hand, Snail silencing diminished the capacity of BMP7 to induce GFAP expression and abrogated the ability of BMP7 to decrease the self-renewal of GSCs (Supplementary Figure S9A, B and ref. [13]). On the other hand, TGFβ1 is well known to induce EMT [27]; in GBM, we uncovered that TGFβ1 induces Snail and Slug expression, the latter in a Snail-dependent manner (Supplementary Figure S9C). However, the ability of TGFβ1 to promote GSC self-renewal is not Snail dependent (Supplementary Figure S9B). Thus Snail organizes extensive and selective crosstalk with the TGFβ and BMP pathways.
TGFβ1 restores stem-like growth in GSCs expressing Snail. a ELDA of U2987MG cells (A-pcDNA and F-Snail) plated in decreasing numbers (200–1 (cell/s)/well) in the presence or absence of 5 ng/ml TGFβ1 for 10 days (n = 2, technical octaplicate). Steeper slopes indicate higher frequencies of colony-forming cells. A table indicates average stem cell frequency for each group. b–d qRT-PCR analysis of mRNA expression in cells stimulated with or without 5 ng/ml TGFβ1 for 72 h (n = 3, technical triplicate). Results are expressed as mean ± SD. Statistical comparison (two-way Anova) indicates significant differences at: *p < 0.05, **p < 0.01, ***p < 0.001
Exogenous TGFβ1 rescues the stem-like features in Snail-expressing GSCs
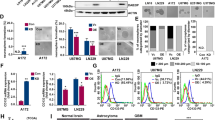
Snail transcriptionally represses the TGFB1 gene and thereby suppresses the pro-stemness effects of this cytokine in GBMs, and promotes efficient GBM astrocytic fate switch. We therefore investigated whether exogenous TGFβ1 added to the GSC microenvironment might bypass the cell-autonomous effects of Snail. By stimulating Snail-expressing GSCs with recombinant TGFβ1, we observed rescue of many phenotypic effects caused by Snail. TGFβ1 treatment restored sphere-forming capacity (Fig. 5a); enhanced TGFB1, inhibitory SMAD6/7, and stem cell marker NESTIN and LIF gene expression; and reduced ID4 and SPARCL1 levels (Fig. 5b, c). However, TGFβ1 did not affect BMP4 or MEF/ELF4 (Fig. 5d); it even had a positive effect on GFAP expression (Fig. 5c).
U2987MG-Snail cells upregulate BMP expression and repress TGFβ1 signaling in order to promote astrocytic fate and repress stemness. We observed that treating GSCs with BMP7 and the TGFβ type I receptor inhibitor LY2157299/galunisertib (L2) partially mimicked the effects of Snail (Fig. 6); we chose BMP7 as it has been widely used to induce GBM cell differentiation [14, 28]. This combinatorial treatment strongly reduced sphere formation by U2987MG cells (Fig. 6a), it induced ID4, and reduced TGFB1 expression (Fig. 6b, c), almost to the levels measured in U2987MG-Snail cells; however, this combination failed to repress stem cell marker, inhibitory SMAD6/7 or BMP4 mRNA levels (Fig. 6b–d). As an inverse pharmacological control, we tested the GSC response to the BMP receptor inhibitor DMH1 in combination with TGFβ1 ligand. In agreement with all previous results, DMH1 enhanced the response to TGFβ1 in terms of reducing BMP4, ID4, and SPARCL1 expression; however, GFAP expression could not be reduced, on the contrary, it was further increased (Fig. 6b–d). Moreover, DMH1/TGFβ1 co-treatment recovered the expression of endogenous TGFβ1 and rescued inhibitory SMAD6/7 and stem cell marker NESTIN, LIF, and MEF expression in U2987MG-Snail cells (Fig. 6b–d).
BMP7 and TGFβ inhibitor co-treatment partially phenocopies Snail function in GSCs. a ELDA of U2987MG cells (A-pcDNA and F-Snail) plated in decreasing numbers (200–1 (cell/s)/well). A-pcDNA3 cells were treated with or without 30 ng/ml BMP7 in the presence or absence of TGFβ inhibitor LY2157299 (L2, 2 µM). F-Snail cells were treated with or without 5 ng/ml TGFβ1 in the presence or absence of DMH1 (0.5 µM). Spheres were counted on day 10 (n = 2, technical octaplicate); steeper slopes indicate higher frequencies of colony-forming cells. The table indicates average stem cell frequency for each group. b–d qRT-PCR analysis of mRNA expression in A-pcDNA cells treated with BMP7 (30 ng/ml) and TGFβ inhibitor (L2, 2 µM) and in F cells treated with TGFβ1 (5 ng/ml) and BMP inhibitor (DMH1, 0.5 µM) for 72 h (n = 2, technical triplicate). Results are expressed as mean ± SD. Statistical comparison (one-way Anova) indicates significant differences at: *p < 0.05, **p < 0.01, ***p < 0.001
We investigated whether the in vitro effects of DMH1/TGFβ1 or L2/BMP7 co-treatment were persistent in vivo. We marked GBM cells with the green fluorescent protein (GFP), generating stably transfected U2987MG-GFP and U2987MG-Snail-GFP clones; these were then treated with the different factors for 72 h, prior to orthotopic injection in the brains of newborn mice. Macroscopic tumors were observed only in mice injected with U2987MG cells and unexpectedly even in mice injected with cells pretreated with BMP7 (Fig. 7a). U2987MG-Snail cells formed only microscopic tumors, independently of the pretreatment applied (Supplementary Figure S10, GFP staining). In agreement with the incidence of macroscopic tumors, U2987MG cells were more proliferative compared to U2987MG-Snail cells in the tumor tissue (Fig. 7b, c, Ki67 staining) and TGFβ1 inhibition (L2) reduced proliferation, whereas BMP7 pretreatment did not; it even slightly enhanced Ki67 staining in U2987MG parental cells. Pretreatment with TGFβ1 or BMP receptor inhibitor (DMH1) separately promoted U2987MG-Snail tumor cell proliferation (Fig. 7b, c). The stem cell marker Nestin was expressed in vivo at considerably lower levels in U2987MG-Snail cells compared to U2987MG tumors as expected by the expression profile in vitro; TGFβ1 (L2) and BMP (DMH1) receptor inhibitors enhanced Nestin expression in vivo, as they did in vitro (Fig. 7b, c, Nestin staining). ID4, which marks BMP signaling activity, was expressed at the same level in both U2987MG and U2987MG-Snail tumors; BMP7 pretreatment, as expected, enhanced ID4 expression (Supplementary Figure S11). We monitored phospho-SMAD2 and PAI1/SERPINE1 in order to assess TGFβ signaling activity in these tumors (Fig. 7b, c, Supplementary Figure S11). In U2987MG-Snail cells, both proteins gave weaker signals, and these signals were enhanced after TGFβ or DMH1 inhibitor pretreatment, as expected. These results show that Snail provides dual and opposite regulation of BMP and TGFβ pathways, in order to favor astrocytic fate and repress stemness (Supplementary Figure S12). By regulating TGFβ family ligand expression, Snail can suppress GBM tumorigenesis.
Snail suppresses GBM in vivo. a Tumor formation in orthotopically xenografted immunodeficient mice. b Immunohistochemistry of GFP, Ki67, Nestin, and phospho-SMAD2. c Immunohistochemistry quantification: (left) % Ki67-positive nuclei expressed as a percentage of tumor area; (center and right) reciprocal intensity of Nestin and phosho-SMAD2. Data show mean ± SD of 9–20 fields of 3 mice. Statistical comparison (one-way Anova) indicates significant differences at: *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
Snail is involved in the progression of epithelial cancers but is less studied in brain tumors. In epithelial tumors, it is well established that expression of Snail correlates with EMT, invasion, gain of cancer stem cell properties, and radio- and chemo-resistance [17]. Snail-induced EMT also contributes to tissue fibrosis [17, 29]. In glioma, Snail promotes cell migration [18, 19], as GBMs expressing high Snail are more invasive compared to GBMs expressing low Snail levels [13]. However, mice in which Snail-expressing cells had been injected intracranially showed longer survival and reduced efficiency of tumor formation [13]. These results indicate that Snail promotes invasion and at the same time limits GBM growth. We have now elucidated a novel mechanism by which Snail exerts tumor-suppressor effects in GBM and which explain how Snail can lead to reduced tumor burden. Snail enhances BMP signaling (Figs. 1 and 2), which in turn primes an astrocytic fate switch, in agreement with the differentiation potential of BMPs in GBMs [11,12,13,14]. Blocking the BMP pathway using a BMP type I receptor kinase small molecule inhibitor (DMH1), knockdown of endogenous BMP receptors or SMAD1/5 and extracellular ligand trapping using a physiological BMP antagonist (Noggin) abrogated the capacity of Snail to promote astrocytic fate (Fig. 2; Supplementary Figure S4, 5). In contrast, endogenous activation of BMP signaling did not contribute to the repression of stem cell markers by Snail in the GSCs. This finding distinguishes differentiation mechanisms mediated by Snail from mechanisms controlling stemness in GSCs.
A key factor that links Snail to suppression of GSC stemness is TGFβ1 itself (Fig. 3). Snail represses the TGFB1 gene, a well-known tumor promoter in GBM, which enhances GSC proliferation and self-renewal in addition to promoting intratumoral angiogenesis [8, 10, 30]. Thus, Snail promotes BMP signaling and subsequent cell differentiation, and by suppressing TGFβ1 and its downstream signaling it also suppresses the survival and self-renewal of GSCs. Accordingly, and in agreement with previous reports [8, 10, 30], inhibiting TGFβ1 signaling reduces GBM proliferation in vivo (Fig. 7). In this GBM model, BMP7 pretreatment was not sufficient to suppress tumor development, possibly due to the dose and length of the treatment applied (Fig. 7). This finding also underscores that BMPs may not induce terminal differentiation of tumor cells but rather promote a plastic switch in cell fate [11]. Finally, TGFβ1 pretreatment enhanced tumor cell proliferation, stemness, and sustained TGFβ signaling in vivo (Fig. 7), confirming its pro-tumorigenic action in glioblastoma [8,9,10]. Moreover, blocking the BMP pathway in U2987MG-Snail cells phenocopied TGFβ pretreatment in vivo (Fig. 7), confirming the importance of Snail-induced BMP signaling to prime for an astrocytic fate and maintain tumor-suppressor effects.
Mechanistically, our work places Snail as a direct regulator of TGFβ1 expression, as Snail binds to and represses the TGFB1 promoter (Figs. 3 and 4). Snail can activate TGFβ signaling in mammary and renal epithelial cells [29, 31]; however, direct binding of Snail to the TGFB1 promoter was not shown previously. Moreover, we now show that Snail, via its N- and C-terminal domains, interacts with the BMP and TGFβ R-SMAD MH1-Linker domains (Fig. 4; Supplementary Figure S7, 8).
The antagonistic effects of BMP signaling against TGFβ are well established in embryonic development and in cancer [27]. It remains to be examined further whether BMPs promote SMAD1/5 complexes with Snail to regulate additional target genes, which are important for astrocytic differentiation and suppression of GSC stemness. Repression of TGFB1 by Snail is a key event that controls GBM stemness; TGFβ1 alone or in combination with the BMP type I receptor inhibitor, DMH1, helped GSCs recover their self-renewal capacity and the expression of specific stemness genes, such as NESTIN and LIF (Figs. 5 and 6). Combining exogenous TGFβ1 with DMH1 also rescued inhibitory SMAD6/7 expression and blocked BMP4, ID4, and astrocytic marker SPARCL1 expression (Fig. 6). Interestingly, the combination of exogenous TGFβ1 with DMH1 did not block GFAP expression (Fig. 6) as GFAP can be induced by TGFβ1 in astrocytes during wound repair mechanisms in the central nervous system [32, 33]. Altogether, our data show that Snail limits the tumorigenic potential of GBM by blocking GSC self-renewal and promoting a fate switch toward the astrocytic lineage by controlling in opposite ways different arms of the TGFβ family, inducing the BMP arm, and repressing the TGFβ1 arm (Supplementary Figure S12). This mechanism underscores the importance of sequential and interdependent loops of signaling by distinct TGFβ family members, a phenomenon of widespread importance in cancer progression [27], which is now shown to partake the key transcriptional regulator Snail.
Materials and methods
Reagents
The mammalian expression constructs pcDNA3, pEGFP-N1, and pcDNA3-Flag-tagged SMAD2/3/4 have been described [34]. The pcDNA3-Flag-SMAD1/5 vectors were kindly provided by K. Miyazono and T. Imamura (Tokyo University and Ehime University, Japan, respectively) [35]. pGEX vectors encoding GST-SMAD1 and its deletion mutants have been described [36]. The human Snail domain plasmids were provided by A. García de Herreros (IMIM, Barcelona, Spain) [37]. The silencing vectors pSuperior-Neo-GFP (pSGN/shControl) and shSnail or pcDNA3-Snail are described [34].
Cell culture and transfection
Human embryonic kidney 293T cells and GBM cells U2987MG (previously described [38]) were cultured in Dulbecco’s modified Eagle’s medium or Minimum Essential Medium (MEM), respectively, supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Both cell lines scored negatively for mycoplasma infection; the 293T cells were authenticated by Short Tandem Repeat profiling. U2987MG cells expressing pcDNA3, pcDNA3-Snail, pSGN, or shSnail have been described [13]. Prior to orthotopic brain injection, cells were further transfected stably with the pEGFP-N1 plasmid using the FUGENE-HD (Promega) protocol, generating U2987MG-GFP and U2987MG-Snail-GFP clones, respectively.
GBM cells in MEM plus 10% FBS or stem cell media (see extreme limiting dilution assay (ELDA) protocol) were treated with TGFβ1 (5 ng/ml, Peprotech), BMP7 (30 ng/ml, a gift from K. Sampath, Sanofi-Genzyme Research Center, Framingham, USA), Noggin (0.25 µg/ml, R&D Systems), TGFβ type I receptor kinase inhibitor LY2157299 (L2, 2.5 µM, Sigma-Aldrich), and BMP type I receptor kinase inhibitor DMH1 (500 nM, synthesized by the Ludwig Cancer Research Ltd). Dimethyl sulfoxide was the vehicle for the chemical inhibitors and 1 mM HCl/0.1% bovine serum albumin for the growth factors.
siRNA transfection
Cells were transfected at 80% confluence using a mixture of DharmaFECT 1 and siRNA (20 nM) twice on 2 consecutive days, according to the manufacturer (Dharmacon/GE Healthcare). The human-specific siRNA oligonucleotides were: ON-TARGETplus-SMARTpool-siCONTROL (D-001810-10); ON-TARGETplus-SMARTpool-SMAD1 (L-012723); ON-TARGETplus-SMARTpool-SMAD5 (L-015791); ON-TARGETplus-SMARTpool-ACVR1 (L-004924); ON-TARGETplus-SMARTpool-BMPR1B (L-004934), and ON-TARGETplus-SMARTpool-SNAI1 (L-010847).
RNA extraction and expression analysis
Cellular RNA was purified with the Nucleospin RNA Kit (Macherey-Nagel) or Qiagen RNeasy Mini Kit (Qiagen); gene expression was analyzed by real-time PCR as described [39] with specific primers (Supplementary Table I). TaqMan® Array Human TGFB Pathway (ThermoFisher Scientific) analysis followed the manufacturer’s instructions on the ABI 7900 HT instrument (Applied Biosystems, California, USA).
The Affymetrix Genechip system U133 plus 2.0 and the GeneChip 3’IVT Express protocol were used. Microarray hybridization, scanning, and data normalization was performed by the Uppsala University Array Platform according to standard procedures (http://www.medsci.uu.se/klinfarm/arrayplatform/). Two independent samples for each cell clone were analyzed, and the replicate is denoted as “bis”. To identify genes that were significantly regulated by Snail, we compared U2987MG-Snail to U2987MG-mock cells and filtered gene expression based on an adjusted p < 0.01 using geWorkbench [40]. The U2987MG expression profiles have been deposited to Array Express https://www.ebi.ac.uk/arrayexpress/ with accession number E-MTAB-6151.
Immunoblotting and coimmunoprecipitation
Total proteins from transfected 293T or stimulated U2987MG cells were extracted in 0.5 % Triton X-100, 0.5 % sodium deoxycolate, 20 mM Tris pH 7.4, 150 mM NaCl, 10 mM EDTA and complete protease inhibitor cocktail (Roche) during 30 min on ice and centrifuged at 13,000 r.p.m. for 10 min at 4 °C to remove insoluble debris. Proteins were analyzed by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) or immunoprecipitated with anti-Flag (1 µg), anti-SMAD1 (1 µg), anti-SMAD4 (1 µg), anti-SMAD2/3 (2 µg), or anti-Snail (3 µg) antibodies, and after four washes with lysis buffer, immunocomplexes were resolved by SDS-PAGE and detected by immunoblotting. The antibodies used were: rabbit-anti-Nestin (Millipore, MAB5326), rabbit-anti-phospho-SMAD2 (home-made) [34], rabbit-anti-SMAD3 (9523), rabbit-anti-phospho-SMAD1/5 (9511), rabbit-anti-SMAD5 (9517), and rabbit-anti-Snail (C15D3) (Cell Signaling); mouse-anti-SMAD1 (ab53745, Abcam); rabbit-anti-GFAP (sc-6171), mouse-anti-SMAD4 (sc-7966), and mouse-anti-GST (sc-138) (Santa Cruz Inc.); mouse-anti-SMAD2 (EB05147, Everest Biotech); mouse-anti-SMAD2/3 (610843, BD Biosciences); mouse-anti-HA (11666606001, Roche); rabbit-anti-GFP (A111222, Invitrogen); and mouse-anti-Flag M5 (F4042, Sigma). Mouse-anti-GAPDH (AM4300, Ambion) was used as loading control in immunoblot analyses.
Chromatin immunoprecipitation
U2987MG (107) cells were crosslinked with 1% formaldehyde, neutralized with 0.125 M glycine, washed with ice-cold phosphate-buffered saline (PBS), and resuspended in 50 mM Tris-HCl, pH 8.1, 1% SDS, 10 mM EDTA, and complete protease inhibitors. ChIP assay was as described previously [39]. The primers for the human TGFB1 promoter were forward: GGGTGTTGAGTGACAGGAGG and reverse: GAGGGTCTGTCAACATGGGG.
GST pulldown
GST-pulldown assays were as described previously [36]. Interaction assays of GST-SMAD1 with endogenous U2987MG Snail used cell extracts prepared as in immunoblot with additional 1 mM NaOVO3. Extract aliquots corresponding to ~107 cells were incubated with glutathione-sepharose beads carrying 2.5 μg of GST-SMAD fusions overnight at 4 °C. Bound proteins washed with lysis buffer were resolved by 8% SDS-PAGE.
Immunofluorescence microscopy
Immunofluorescence protocol has been described previously [13]. The pSMAD1/5, SMAD2, pSMAD2, and SMAD3 antibodies used were as described under immunoblotting. Random (5–10) pictures were taken with a Nikon Eclipse 90i microscope (20× objective) at the same exposure time for each protein. ImageJ64 10.2 software (National Institutes of Health, Bethesda, MD, USA) was used to quantify pixels in the nuclear area.
Extreme limiting dilution assay
ELDA was carried out as described [41]. U2987MG cells were grown in 96-well plates in decreasing numbers (200–1 (cell/s)/well) in 200 μl of serum-free MEM supplemented with 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 25 ng/ml epidermal growth factor, 25 ng/ml basic fibroblast growth factor, and B27 complement. On day 10, the number of wells containing spheres for each cell plating density was recorded and plotted using the online ELDA analysis program (http://bioinf.wehi.edu.au/software/elda), which estimates a stem cell frequency for each condition [41]. The data are plotted as log fraction of wells without spheres as a function of the plated cell number.
EV profile analysis
Cells were extensively washed with PBS and incubated for several hours in serum-free medium to eliminate vesicles stemming from serum. The wash step was repeated before adding fresh serum-free medium and incubating for 48 h, when media was sequentially centrifuged at 3000 × g for 5 min to pellet cells and at 10,000 × g for 10 min to remove cell debris. Supernatants were filtered (0.45 μm PES filter, VWR) and pelleted at 100,000 × g for 2 h at 4 °C. EVs were resuspended in PBS containing protease inhibitors (Roche) and lysed in (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, pH 8, 1% Triton X-100, and 0.1% sodium deoxycholate) and protein concentration was measured by the Dot-it-Spot-it Kit (http://dot-it-spot-it.com) according to the manufacturer’s protocol. Specific proteins were analyzed by multiplex PEA that is a 96-plex immunoassay for protein detection in 1 µl of sample as described previously [42]. Normalized protein expression (log2-scale) was calculated as described previously [43].
Enzyme-linked immunosorbent assay (ELISA)
U2987MG conditioned media were concentrated 50× through Amicon Ultra-15 centrifugal filters (Millipore) at 3000 × g for 15 min at 4 °C. Secreted BMP4 and TGFβ1 were measured using the human BMP4-ELISA Kit and the human TGFβ1-Duoset ELISA Kit, respectively (R&D Systems), according to the manufacturer’s instructions.
Animal experiments
Animal experiments were performed in accordance to the regulations of Uppsala University and approved by the local ethics committee. U2987MG-pcDNA3-GFP and U2987-MG-Snail-GFP (105 cells in 2 µl PBS) were stereotactically injected into neonatal NOD-SCID mice (P0–2, both genders used) using a motorized stereotactic injector (Stoelting) at a pulse rate of 1.4 µl/min. The stereotactic coordinates, measured from lambda, were: anterio-posterior 1.5 mm, meso-lateral 0.7 mm, and dorso-ventral 1.5 mm. Mice were euthanized upon signs of sickness or distress, otherwise, at 14 weeks postinjection in compliance with the ethical guidelines. Mouse brains were fixed in 4% formalin and tumors were examined histologically. Animal numbers are reported in the figures; gender was not considered as a parameter neither did any randomization method.
Immunohistochemistry (IHC)
Paraffin embedding of formalin-fixed samples, sectioning, IHC, and slide scanning were performed by the Science for Life Laboratory facilities at the Department of Immunology, Genetics, and Pathology of Uppsala University, Sweden, as described [44]. Primary antibodies diluted in UltraAb Diluent (Thermo Fisher Scientific) were applied for 30 min at room temperature. The antibodies used at the indicated dilutions were: anti-GFP (Invitrogen A111222, 1:2000), anti-Ki67 (Abcam Ab15580, 1:1000), anti-Nestin (Millipore MAB5326, 1:100), anti-ID4 (Santa Cruz, sc-491, 1:100), anti-phospho-SMAD2 (Invitrogen 44-244 G, 1:200), and anti-PAI1 (Abcam, Ab66705 1:200). The slides were incubated with secondary antibody (Abcam Ab6721; Goat-Anti-Rabbit IgG H&L (horseradish peroxidase), 1:1000) for 30 min at room temperature and scanned using the automated scanning system Aperio XT (Aperio Technologies). Chromogen intensity was quantified using the reciprocal intensity (r) method [45], where: r = 255−y, 255 being the maximum pixel intensity of unstained area (as measured by the standard intensity function in the Fiji software) and y = mean intensity of the selected tumor area. Reciprocal intensity was quantified from one to six fields of three mice per group; the number of fields depended on the tumor size, and all analyses were performed without any blinding.
Statistical analysis
For each experimental technique, unless otherwise stated in the methods or figure legends, individual experiments were repeated three or more times; this allowed us to reach high statistical significance in all the reported assays. Data were analyzed using Prism GraphPad v6.0. A two-tailed Student's t-test was performed in two-group comparisons. One-way analysis of variance (Anova), followed by Sidak’s multiple comparison test, was performed in multiple group comparisons. Two-way Anova was used when comparing TGFβ1 treatment on two different cell populations (pcDNA3 versus pcDNA3-Snail). The variances were similar between the groups under comparison in all these cases.
References
Yan K, Yang K, Rich JN. The evolving landscape of glioblastoma stem cells. Curr Opin Neurol. 2013;26:701–7.
Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 2011;71:4055–60.
Haar CP, Hebbar P, Wallace GC, Das A, Vandergrift WA, Smith JA, et al. Drug resistance in glioblastoma: a mini review. Neurochem Res. 2012;37:1192–200.
Chen J, Li Y, Yu T-SS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6.
Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702.
Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73.
Sakaki-Yumoto M, Katsuno Y, Derynck R. TGF-β family signaling in stem cells. Biochim Biophys Acta. 2013;1830:2280–96.
Bruna A, Darken RS, Rojo F, Ocaña A, Peñuelas S, Arias A, et al. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–60.
Peñuelas S, Anido J, Prieto-Sánchez RM, Folch G, Barba I, Cuartas I, et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–27.
Ikushima H, Todo T, Ino Y, Takahashi M, Saito N, Miyazawa K, et al. Glioma-initiating cells retain their tumorigenicity through integration of the sox axis and oct4 protein. J Biol Chem. 2011;286:41434–41.
Carén H, Stricker SH, Bulstrode H, Gagrica S, Johnstone E, Bartlett TE, et al. Glioblastoma stem cells respond to differentiation cues but fail to undergo commitment and terminal Cell-Cycle arrest. Stem Cell Rep. 2015;5:829–42.
Piccirillo SGM, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–5.
Savary K, Caglayan D, Caja L, Tzavlaki K, Bin Nayeem S, Bergström T, et al. Snail depletes the tumorigenic potential of glioblastoma. Oncogene. 2013;32:5409–20.
Tate CM, Pallini R, Ricci-Vitiani L, Dowless M, Shiyanova T, D’Alessandris GQ, et al. A BMP7 variant inhibits the tumorigenic potential of glioblastoma stem-like cells. Cell Death Differ. 2012;19:1644–54.
Yan K, Wu Q, Yan DH, Lee CH, Rahim N, Tritschler I, et al. Glioma cancer stem cells secrete Gremlin1 to promote their maintenance within the tumor hierarchy. Genes Dev. 2014;28:1085–100.
Herbertz S, Sawyer JS, Stauber AJ, Gueorguieva I, Driscoll KE, Estrem ST, et al. Clinical development of galunisertib (LY2157299 monohydrate), a small molecule inhibitor of transforming growth factor-beta signaling pathway. Drug Des Dev Ther. 2015;9:4479–99.
Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850.
Mahabir R, Tanino M, Elmansuri A, Wang L, Kimura T, Itoh T, et al. Sustained elevation of Snail promotes glial-mesenchymal transition after irradiation in malignant glioma. Neuro Oncol. 2014;16:671–85.
Yang HW, Menon LG, Black PM, Carroll RS, Johnson MD. SNAI2/Slug promotes growth and invasion in human gliomas. BMC Cancer. 2010;10:301.
Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–78.
Kobayashi A, Okuda H, Xing F, Pandey PR, Watabe M, Hirota S, et al. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J Exp Med. 2011;208:2641–55.
Miyazono K, Miyazawa K. Id: a target of BMP signaling. Sci STKE 2002;2002:pe40.
Whissell G, Montagni E, Martinelli P, Hernando-Momblona X, Sevillano M, Jung P, et al. The transcription factor GATA6 enables self-renewal of colon adenoma stem cells by repressing BMP gene expression. Nat Cell Biol. 2014;16:695–707.
Caja L, Bellomo C, Moustakas A. Transforming growth factor β and bone morphogenetic protein actions in brain tumors. FEBS Lett. 2015;589:1588–97.
Marinescu VD, Kohane IS, Riva A. The MAPPER database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res. 2005;33:D91–7.
Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol. 2009;11:943–50.
Caja L, Kahata K, Moustakas A. Context-dependent action of transforming growth factor beta family members on normal and cancer stem cells. Curr Pharm Des. 2012;18:4072–86.
González-Gómez P, Crecente-Campo J, Zahonero C, de la Fuente M, Hernández-Laín A, Mira H, et al. Controlled release microspheres loaded with BMP7 suppress primary tumors from human glioblastoma. Oncotarget. 2015;6:10950–63.
Grande MT, Sánchez-Laorden B, López-Blau C, De Frutos CA, Boutet A, Arévalo M, et al. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat Med. 2015;21:989–97.
Tchaicha JH, Reyes SB, Shin J, Hossain MG, Lang FF, McCarty JH. Glioblastoma angiogenesis and tumor cell invasiveness are differentially regulated by β8 integrin. Cancer Res. 2011;71:6371–81.
Dhasarathy A, Phadke D, Mav D, Shah RR, Wade PA. The transcription factors Snail and Slug activate the transforming growth factor-beta signaling pathway in breast cancer. PLoS ONE. 2011;6:e26514.
Doyle KP, Cekanaviciute E, Mamer LE, Buckwalter MS. TGFβ signaling in the brain increases with aging and signals to astrocytes and innate immune cells in the weeks after stroke. J Neuroinflamm. 2010;7:62.
Schachtrup C, Ryu JK, Helmrick MJ, Vagena E, Galanakis DK, Degen JL, et al. Fibrinogen triggers astrocyte scar formation by promoting the availability of active TGF-beta after vascular damage. J Neurosci. 2010;30:5843–54.
Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008;283:33437–46.
Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, et al. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–80.
Watanabe Y, Papoutsoglou P, Maturi V, Tsubakihara Y, Hottiger MO, Heldin C-H, et al. Regulation of bone morphogenetic protein signaling by ADP-ribosylation. J Biol Chem. 2016;291:12706–23.
Villagrasa P, Díaz VM, Viñas-Castells R, Peiró S, Del Valle-Pérez B, Dave N, et al. Akt2 interacts with Snail1 in the E-cadherin promoter. Oncogene. 2012;31:4022–33.
Ferletta M, Caglayan D, Mokvist L, Jiang Y, Kastemar M, Uhrbom L, et al. Forced expression of Sox21 inhibits Sox2 and induces apoptosis in human glioma cells. Int J Cancer. 2011;129:45–60.
Tan E-J, Kahata K, Idas O, Thuault S, Heldin C-H, Moustakas A. The high mobility group A2 protein epigenetically silences the Cdh1 gene during epithelial-to-mesenchymal transition. Nucleic Acids Res. 2015;43:162–78.
Floratos A, Smith K, Ji Z, Watkinson J, Califano A. geWorkbench: an open source platform for integrative genomics. Bioinformatics. 2010;26:1779–80.
Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8.
Larssen P, Wik L, Czarnewski P, Eldh M, Löf L, Ronquist KG, et al. Tracing cellular origin of human exosomes using multiplex proximity extensionassays. Mol Cell Proteom. 2017;16:502–11.
Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9:e95192.
Kampf C, Olsson I, Ryberg U, Sjöstedt E, Pontén F. Production of tissue microarrays, immunohistochemistry staining and digitalization within the human protein atlas. J Vis Exp. 2012:e3620.
Nguyen D, Zhou T, Shu J, Mao J Quantifying chromogen intensity in immunohistochemistry via reciprocal intensity. Cancer InCytes. 2013;2. https://doi.org/10.1038/protex.2013.097.
Acknowledgements
We thank Bengt Westermark and Karin Forsberg-Nilsson (Dep. Immunology, Genetics Pathology, Uppsala University) for human GBM cells; Kuber Sampath (Sanofi-Genzyme Research Center, Framingham) for recombinant BMP7; Kohei Miyazono (Tokyo University) and Takeshi Imamura (Ehime University) for SMAD vectors; Antonio Garcia de Herreros (IMIM, Barcelona) for Snail vectors; Per-Henrik Edqvist (Dep. Immunology, Genetics, Pathology, Uppsala University) and SciLifeLab Tissue Profiling platform for IHC analysis; and Hanna Göransson Kultima (Dep. Medical Sciences, Uppsala University) and Bioinformatics Infrastructure for Life Sciences, Uppsala University for assistance with microarray analysis. We thank George Divolis and members of our group for suggestions and discussions.
Funding
This work was supported by: Ludwig Cancer Research; Swedish Cancer Society (CAN 2012/438, CAN 2015/438 to ArM); and Swedish Research Council (K2013-66X-14936-10-5 to ArM, 2015-02757 to C-HH). LC was supported by postdoctoral fellowships from Lisa Erikssons minnesfond (2013) and Swedish Cancer Society (CAN 2012/1186).
Author contributions
Conception and design: LC, ArM. Data acquisition: LC, KT, MSD, E-JT, GH, NPM, AnM, LW, YW, KS. Data analysis and interpretation: LC, KT, MSD, MK-M, LU, C-HH, ArM. Article drafting and critical revision for important intellectual content: LC, C-HH, ArM. Final approval prior to publication: all authors.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Caja, L., Tzavlaki, K., Dadras, M.S. et al. Snail regulates BMP and TGFβ pathways to control the differentiation status of glioma-initiating cells. Oncogene 37, 2515–2531 (2018). https://doi.org/10.1038/s41388-018-0136-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0136-0
This article is cited by
-
The long non-coding RNA LINC00707 interacts with Smad proteins to regulate TGFβ signaling and cancer cell invasion
Cell Communication and Signaling (2023)
-
BMP signaling in cancer stemness and differentiation
Cell Regeneration (2023)
-
Identification of ligand–receptor pairs that drive human astrocyte development
Nature Neuroscience (2023)
-
Competition between type I activin and BMP receptors for binding to ACVR2A regulates signaling to distinct Smad pathways
BMC Biology (2022)
-
Structures and biological functions of zinc finger proteins and their roles in hepatocellular carcinoma
Biomarker Research (2022)