Abstract
Poorly differentiated colorectal cancers (CRCs) are more aggressive and lack targeted therapies. We and others previously reported the predominant role of tumor-suppressor NDRG2 in promoting CRC differentiation, but the underlying mechanism is largely unknown. Herein, we demonstrate that NDRG2 induction of CRC cell differentiation is dependent on the repression of E3 ligase Skp2 activity. In patients and Ndrg2 knockout mice, NDRG2 and Skp2 are negatively correlated and associated with cell differentiation stage. Further, NDRG2 suppression of Skp2 contributes to the inductions and stabilizations of p21 and p27, which are Skp2 target proteins for degradation. The reduction of either p21 or p27 levels by shRNA can decrease NDRG2-induced AKP activity and resume cell growth inhibition, thus both p21 and p27 are required for NDRG2 effect on the promotion of cell differentiation in CRCs. The mechanistic study shows that NDRG2 suppresses β-catenin nuclear translocation and decreases the occupancy of β-catenin/TCF complex on Skp2 promoter, potentially through dephosphorylating GSK-3β. By subjecting a series of NDRG2 deletion mutants to Skp2 expression, the loss of NH2-terminal domain can completely abolish NDRG2-dependent differentiation induction. Supporting the biological significance of the reciprocal relationship between NDRG2 and Skp2, an NDRG2low/Skp2high gene expression signature correlates with poor CRC patient outcome and could be considered as a diagnostic marker of CRCs.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is commonly diagnosed. Differentiation deficiency is one of the key characteristics. Poorly differentiated CRC shows high proliferation and metastasis capacity, which has a serious impact on patient survival and prognosis [1]. Molecularly, the vast majority of poorly differentiated CRCs contain constitutive WNT/β-catenin signal activation. Due to the activated WNT signaling mutations in adenomatous polyposis coli (APC) or axin/conductin, nuclear translocation of β-catenin is induced, and consequently contributes to cell fate determination via β-catenin/transcription factor (TCF) complexes [2,3,4,5]. Free cytoplasmic β-catenin is destabilized by a multiprotein complex containing Axin, glycogen synthesis kinase 3β (GSK-3β) and the APC tumor suppressor. GSK-3β phosphorylates β-catenin, which earmarks β-catenin for ubiquitination and degradation by the SKP1-cullin-F box protein (SCF) complex [6, 7]. The GSK-3β inactivation by APC mutation or oncogenic phosphatidylinositol 3 kinase (PI3K)/AKT activation leads to the β-catenin/TCF complex formation, and further induced TCF target gene expression levels, such as Myc, cyclin D1 [8, 9].
Another character of cellular differentiation is the diminished rate of cell cycle progression. The G1-S transition is driven mainly by cyclin-dependent kinase (CDK) 2, which is controlled by CDK inhibitor p21WAF1 (p21) and p27KIP1 (p27) [10, 11]. Stabilities of intracellular p21 and p27 are predominantly regulated by S-phase kinase-associated protein 2 (Skp2) [12, 13]. Based on the key role of Skp2 as an E3 ligase in cancer development, the related oncogenic pathways have been investigated, such as PI3K/AKT pathway, WNT/β-catenin pathway [14,15,16]. Especially, Skp2 is shown transcriptionally regulated by β-catenin/TCF complex through recognition of two TCF/lymphoid enhancer-binding factor 1 (LEF1) consensus binding sites on Skp2 promoter [16].
N-Myc downstream-regulated gene 2 (NDRG2) as a member of the NDRG family was first identified in our laboratory [17]. As a tumor suppressor, NDRG2 has been found downregulated in multiple tumors including CRCs [18]. Notably, our recent work demonstrated that NDRG2 is involved in the metabolic reprogramming [19], and TGF-β-induced epithelial–mesenchymal transition [20] in CRCs. Accumulating evidence indicates that NDRG2 is involved in regulating cell differentiation and proliferation. NDRG2 levels are positively correlated with tumor differentiation stages in various cancers [21, 22]. Elevated NDRG2 expression confers to the induction of E-cadherin expression and prolongs overall survival in CRC patients [21]. Although there is still no direct evidence to prove how NDRG2 coordinates cancer cell proliferation and differentiation, and the underlying mechanism is largely unknown. In this report, we explored the association of NDRG2 with CRC differentiation in patients and transgenic mice, and reported a reciprocal regulatory relationship between NDRG2 and Skp2 that controls cell differentiation and confers to their aggressive growth.
Results
NDRG2 is a differentiation-specific marker in CRCs
To fully address the clinical significance of NDRG2 in CRC patients, we first sought to determine its expression in CRC and adjacent normal colon tissues from 112 patients. Immunohistochemical and statistical analysis showed that NDRG2 was broadly lowly expressed in the aggressive CRCs (Figs. 1a, b), whereas NDRG2 was predominantly highly expressed in well differentiated CRCs (Fig. 1c). We then correlated NDRG2 involved cell differentiation with CRC patient outcome. The poor differentiation status showed a high correlation with decreased survival (Fig. 1d), demonstrating differentiation is a critical marker for the prognosis of CRC patients. Meanwhile, low NDRG2 expression correlated with decreased survival (Fig. 1e). A further study in silico analysis of 383 primary CRC tissues of the multidimensional data set from The Cancer Genome Atlas (TCGA) also revealed that low NDRG2 expression predicts a poor CRC patient outcome (Fig. 1f), which is due to the hypermethylation of NDRG2 promoter (Fig. 1g). Interestingly, low NDRG2 expression showed a most evident correlation with decreased survival in patients whose tumors also had low p21 or p27 expression (Figs. 1h, i), indicating the CDK inhibitors probably be involved in NDRG2 regulation of CRC malignancy. Thus, NDRG2 is a differentiation-specific marker in CRCs and related to prognosis.
NDRG2 is a differentiation-specific marker in CRCs. a Immunohistochemistry staining of NDRG2 in human adjacent normal and cancerous colon tissues from CRC patients. b, c NDRG2 expression in human tissues at different stage b or differentiation status c. Student’s t-test was applied for statistical analyses. d, e Kaplan–Meier plots indicate the clinical outcomes for different CRC differentiation status d or NDRG2 expression patterns e by immunohistochemistry staining in CRCs. f-i Silico analysis of 383 primary CRC tissues of the multidimensional data set from The Cancer Genome Atlas (TCGA) cbioportal data set. f Clinical outcomes for NDRG2 expression pattern were evaluated. g The correlation between the TCGA expression and methylation data for NDRG2 in CRC adenocarcinoma. h, i Clinical outcomes for the gene expression patterns given at the top of each panel. e, f,h, i For NDRG2, CDKN1A and CDKN1B, high expression indicates expression above the mean, calculated across all samples. Conversely, low expression indicates expression below the mean. a-i n indicates the number of patient samples evaluated in each analysis. P-values were calculated using the Mantel–Cox log-rank test. **P < 0.01. ***P<0.001
NDRG2 dictates intestinal epithelial cell differentiation
To further characterize NDRG2 function during cell differentiation, we isolated the whole colon of C57BL/6 mice at the age of 1, 2, or 3 months, respectively, which have differentiation status verified by elevated E-cadherin expression and alkaline phosphatase (AKP) activities (Figs. 2a–c). In accordance, Ndrg2 expression was increased in the colon of 2- and 3-month mice accompanied with the increased expression levels of p21 and p27 (Figs. 2a, b). Due to the positive correlation of NDRG2 with p21 and p27, we propose NDRG2 may regulate cell differentiation through these CDK inhibitors.
NDRG2 dictates intestinal epithelial cell differentiation. a Immunofluorescent staining of E-cadherin, Ndrg2, p21, and p27 in intestinal epithelium from C57BL/6 mice at the age of 1, 2, or 3 months as indicated. b Quantification of fluorescence intensity ImageJ. c AKP activities of intestinal epithelial cells from mice at the indicated age. d Schematic representation of the Ndrg2 targeting vector (top), wild-type locus (WT, middle), targeted allele (Ndrg2-floxed, middle), and null allele (Ndrg2-Null, bottom). Exons are represented by solid rectangles. The targeting vector, which is flanked by 3675 bp of 5′ homology and 2138 bp of 3′ homology, contains a LoxP-FRT-Neo-FRT cassette and the second LoxP sequence in introns 1 and 6, respectively. e–g Immunofluorescent staining and intensity quanification of Ndrg2, p21, and p27, and AKP activity were measured in intestinal epithelium from 2-month WT or NDRG2-/- mice. *P < 0.05
Thus, we further generated a mouse line carrying the Ndrg2 conditional knockout allele (Ndrg2-/-) with exons 2–6 flanked by LoxP sites (Fig. 2d). Female homozygotes (Ndrg2-/-) were crossed to CMV-Cre transgenic male mice, which have ubiquitous Cre activity. The whole colon was then isolated from wild-type (WT) mice or Ndrg2-/- mice. The lengths of colon and villus of Ndrg2-/- mice were longer than those of WT mice, indicating NDRG2 is involved in colon development (data not shown). Immunofluorescence assay showed that Ndrg2 expression levels were completely abolished in knockout mice. Moreover, we found the reductions of p21 and p27, and AKP activity after Ndrg2 depletion (Figs. 2e–g), which confirmed our hypothesis that NDRG2 regulation of colon differentiation is through the inductions of p21 and p27.
NDRG2 diminishes CRC cell growth and facilitates cell differentiation
Based on the data from CRC patients and mouse model, we next sought to determine whether gain of NDRG2 function through overexpression would diminish the growth rate (Supplementary Fig. 1A). Expectedly, ectopic NDRG2 expression markedly diminished the cell growth rate and DNA synthesis in HT29 and HCT116 cells (Supplementary Fig. 1B and 1C). Accordingly, the mice with NDRG2-overexpressing HT29 cells developed tumors more slowly than the control group (Figs. 3a, b). In the related tumor section, the proliferation marker Ki-67 is diminished after NDRG2 overexpression (Supplementary Fig. 1D). Thus, NDRG2 overexpression diminishes cell growth through the blockage of DNA synthesis of CRC cells both in vitro and in vivo.
NDRG2 diminishes CRC cell growth and facilitates cell differentiation. a, b NDRG2 suppresses tumor growth of HT29 cells in nude mice. Mice (five animals per group) were injected s.c. in the right limb with 1 × 107 HT29 cells. Tumors were excised and photographed 21 days after injection a. Tumor size was measured over a 3-week period and tumor volume was calculated by the formula (width2 × length × 0.5) b. c–f Cell differentiation status was measured by transmission electron microscopy c, AKP activity analysis d, e, and E-cadherin mRNA expression in the cells labeled on the top of each pane following a 48-h treatment of 2 mM NaBT. g Identification of NDRG2 protein expression in HT29 and HCT116 cells expressing an NDRG2-specific shRNA with or without 2 mM NaBT treatment. h-j Cell differentiation status was measured by AKP activity analysis h, i and E-cadherin mRNA expression j in indicated CRC cells. Data are expressed as means ± SD (n = 3). *P < 0.05
A further cohort study in 681 CRC patients showed that NDRG2 is highly associated with CRC differentiation status, and predominantly highly expressed in well differentiated CRCs (Supplementary Table 1). To better understand how NDRG2 coordinate cell proliferation and differentiation, we determined the differentiation-related phenotypes in CRCs. In quiescent or serum stimulated HT29 and HCT116 cells, the presence of NDRG2 blocked serum-induced cell cycle progression and caused G1 arrest (Supplementary Fig. 2). Intriguingly, NDRG2 overexpression further induced a regular brush border and tight junction morphology (Fig. 3c), and increased AKP activity following the treatment of sodium butyrate (NaBT), a potent inducer of intestinal differentiation (Figs. 3d, e). Adhesion molecule E-cadherin, as a functional marker of differentiation, is necessary for the maintenance of the epithelial cellular structure in CRCs. We here observed that NDRG2 overexpression remarkably increased E-cadherin expression in the presence of NaBT (Fig. 3f). Thus, NDRG2 expression in CRC cells attenuates cell cycle progression and facilitates cell differentiation. Accordingly, NDRG2 reduction by a specific short hairpin RNA (shRNA) can block NaBT-induced AKP activity and E-cadherin expression (Figs. 3g–j). Therefore, NDRG2 is a necessary factor for NaBT-mediated CRC cell differentiation.
NDRG2 elevates p21 and p27 levels
The cyclins, CDKs, and the CDK inhibitors (e.g., p21 and p27) are important for cell cycle progression, and subsequent differentiation [23]. We found that ectopic NDRG2 expression decreased cyclin D1, CDK2, and CDK4 expression levels accompanied with the increase of p21 and p27 protein levels (Fig. 4a). Thus, we reduced p21 or p27 levels by shRNA-mediated knockdown after NDRG2 overexpression (Figs. 4b, e). p21 reduction markedly decreased NDRG2-induced AKP activity and E-cadherin expression in the presence of NaBT (Figs. 4c, d). In accordance, p27 knockdown also blocked NDRG2 effects on AKP activity and E-cadherin expression in HT29 cells (Figs. 4f, g). Thus, NDRG2 expression regulates CRC cell differentiation potentially through increasing p21 and p27 expression, and consequently diminished the rates of cell growth and cell cycle progression. Confirming this point, either p21 or p27 knockdown accelerated the tumor developments, and resumed the growth inhibition of NDRG2 expression in nude mice (Figs. 4h, i).
NDRG2 elevates p21 and p27 levels. We reduced p21 levels using two-lentivirus expressing p21-specific shRNAs in control or NDRG2-overexpressing HT29 cells. We reduced p27 level using a lentivirus expressing a p27-specific shRNA in control or NDRG2-overexpressing HT29 cells. a, b, e Levels of the indicated proteins were determined by western blotting. c, f AKP activity was determined in each cell population following a 48-h treatment of 2 mM NaBT. d, g E-cadherin mRNA levels were determined by qPCR assay. h, i Mice (four animals per group) were injected s.c. in the right limb with 1 × 107 HT29 cells as indicated. Tumors were excised and photographed 5 weeks after injection, and the tumor weight was recorded h. Tumor size was measured over a 5-week period and tumor volume was calculated by the formula (width2 × length × 0.5) i. Data are expressed as means ± SD (n = 3). *P < 0.05
Skp2 repression is required for NDRG2-driven post-translational regulation of p21 and p27
In contrast to the dramatic NDRG2 induction of p21 and p27 protein levels, ectopic NDRG2 expression failed to induce p27 mRNA expression, and partial induced p21 mRNA expression (Fig. 5a), indicating NDRG2 induction of p21 and p27 is not primarily in transcriptional level. As E3 ligase Skp2 potentially targeting p27 and p21 for degradations, we propose Skp2 may contribute to this axis. We first investigate the reciprocal relationship between NDRG2 and Skp2 during CRC cell differentiation. Long-term post-confluence growth of Caco-2 cells showed regular brush borders and tight junctions, which are structural markers of differentiation (Supplementary Fig. 3). The elevated expression levels of NDRG2, p21, and p27 were observed in differentiated Caco-2 cells accompanied with Skp2 reduction (Fig. 5b). Especially, in a 2-day-post-confluence growth induced early differentiation of HT29 cells, NDRG2, and Skp2 were rapidly upregulated and downregulated, respectively (Fig. 5c). Whereas NDRG2 levels increase just before Skp2 levels began to decline. Thus, NDRG2 may repress Skp2 expression during the early stage of cell differentiation. Supporting this hypothesis, we showed that NDRG2 overexpression decreased Skp2 mRNA and protein levels in both HT29 and Caco-2 cells (Figs. 5d, e).
Skp2 repression is required for NDRG2-driven post-translational regulation of p21 and p27. a p21 and p27 mRNA levels were determined in control or NDRG2-overexpressing HT29 and Caco-2 cells. b, c NDRG2 expression was upregulated after 2-day culture, which is correlated with the increased expression levels of p27 and p21, and decreased expression of Skp2. b Levels of indicated proteins in long-term confluence induced differentiation of Caco-2 and HT29 cells. The differentiation induction of Caco-2 and HT29 was performed as described under “Materials and methods” section. c Levels of indicated proteins under post-confluence growth induced differentiation. HT29 cells were seeded at 80% confluence at day 0 or continued to grow for an additional 1 or 2 days. d, e Skp2 protein d and mRNA e expression in HT29 cells with or without NDRG2 expression. f, g Levels of indicated proteins in HT29 cells with or without NDRG2 expression following 10 μg/ml CHX treatment for indicated time. p27 and p21 expression levels were further quantified by ImageJ and normalized to β-actin to determine the protein degradation rate. h, i NDRG2 attenuated p27 and p21 ubiquitination. HT29 cells were transfected with the indicated constructs together with pcDNA3.1( + )-3 HA-Ub and treated with 25 μmol/L MG-132 for 4 h before lysis. Anti-Flag immunoprecipitates from 1.5 mg aliquots of lysates were resolved by 7.5% SDS–PAGE and analyzed by western blotting with anti-HA antibody (top), or anti-FLAG antibody (middle). Cell lysates were subjected to western blotting with an anti-NDRG2 antibody (bottom). WCL whole-cell lysate
Due to Skp2 E3 ligase function, we then determined NDRG2 effect on the half-lifes of p21 and p27 via blocking protein synthesis with cycloheximide (CHX). NDRG2 was stabilized when highly overexpressed, possibly due to exhaustion of the degradation machinery (Fig. 5f). The enhanced stabilities of p21 and p27 were observed in NDRG2 overexpression HT29 cells following CHX treatment (Figs. 5f, g). Further, we determined p21 and p27 ubiquitylations with the expression of HA-tagged-ubquitin (HA-Ub). NDRG2 overexpression was associated with a reduction in the ubiquitination of p21 and p27 relative to control cells (Figs. 5h, i). Therefore, NDRG2 attenuates Skp2 expression and consequently blocks the ubiquitinations and degradations of p21 and p27 in advance, which confers to the promotion of CRC cell differentiation.
NDRG2 represses Skp2 transcription via the suppression of β-catenin
Due to the central role of WNT pathway in the etiology of CRCs and β-catenin/TCF complexes regulation of Skp2 transcription [2, 4, 24], we determined the involvement of β-catenin in NDRG2-mediated Skp2 suppression. Results from the subcellular fraction revealed that NDRG2 overexpression markedly reduced the total and nucleus fraction of β-catenin (Fig. 6a). Consistently, NDRG2 overexpression reduced Topflash activity (Fig. 6b), and the occupancies of TCF/LEF1 on Skp2 promoter (Fig. 6c). Whereas NDRG2 reduction by shRNA increased TCF/LEF1 enrichment on Skp2 promoter (Fig. 6d). Thus, we conclude that NDRG2 blocks TCF/LEF driven Skp2 transcription via suppressing β-catenin nuclear translocation.
NDRG2 represses Skp2 transcription via the suppression of β-catenin. a NDRG2 overexpression decreased the total and nuclear β-catenin expression. β-Actin and Lamin B served as internal controls. b, c HT29 cells were transfected with pcDNA3.1 or pcDNA3.1-NDRG2-overexpressing construct. b NDRG2 attenuated the induction of TCF/LEF activity following LiCl treatment or Wnt-3a expression. c, d The amount of TCF bound to the Skp2 promoter was determined by ChIP following NDRG2 overexpression c or knockdown d. e Levels of the indicated proteins were determined by western blotting in HT29 cells. f, g Levels of the indicated proteins f or Skp2 mRNA g were determined in HT29 cells treated for 24 h with the indicated concentrations of Chir99021. h Immunofluorescent staining and intensity quanification of p-GSK-3β and SKP2 were performed in intestinal epithelium from 2-month WT or NDRG2-/- mice. i Schematic representation of NDRG2 deletion mutants. The full length of NDRG2 contains NH2-terminal region, NDR domain (containing a putative α/β hydrolase fold), and carboxyl-terminal region. NDRG2 deletion mutants without NH2-terminal half (ΔN) or carboxyl-terminal half (ΔC) were generated as indicated. All constructs were tagged with FLAG. j, k We overexpressed full-length NDRG2, NDRG2ΔN, and NDRG2ΔC using relative expressing lentivirus in HT29 cells. The AKP activity i and indicated proteins j were determined, respectively
β-Catenin stability is known regulated by GSK-3β kinase activity. As the NDRG2 was shown decreases PI3K/AKT activity through the regulation of phosphatase and tensin homolog deleted on chromosome ten (PTEN) phosphatase activity [25], it is reasonable to assume that GSK-3β kinase activity is affected by NDRG2 suppression of PI3K/AKT pathway. Expectedly, NDRG2 overexpression diminished the AKT phosphorylation at Ser473 site, and further increased GSK-3β activity via the reduction of GSK-3β phosphorylation at Ser9 site (Fig. 6e). Further, we introduced GSK-3β inhibitor LiCl and Chir99021. The Topflash activity was increased following LiCl treatment or WNT-3a expression, whereas the effect was blocked by NDRG2 overexpression (Fig. 6b). Chir99021 treatment induced Skp2 expression in a dose-dependent manner, and restored Skp2 mRNA and protein levels repressed by NDRG2 overexpression (Figs. 6f, g). Accordingly, the levels of Skp2 and phosphorylated GSK-3β, and the enrichment of TCF/LEF1 on Skp2 promoter were both increased in the colon tissue of Ndrg2 knockout mice (Fig. 6h), indicating the way of NDRG2 repressing Skp2 by WNT pathway comprehensively exists in both human and mouse. By subjecting a series of NDRG2 deletion mutants to Skp2 expression, the loss of NH2-terminal domain can abolish NDRG2 repression of Skp2 and inductions of AKP activity, and p21 and p27 expression levels (Figs. 6i-k), indicating the necessary of NH2-terminal domain for NDRG2-induced CRC differentiation. Therefore, we here demonstrate that NDRG2 suppresses β-catenin expression and nuclear translocation via the regulation of PI3K/AKT/GSK-3β axis, and consequently blocks β-catenin/TCF/LEF complex transcriptional activity on Skp2 promoter.
NDRG2 suppression of Skp2 controls cell differentiation
Given the broad spectrum of biochemical and protooncogenic functions of Skp2, we then sought to determine whether the dramatic alteration of Skp2 levels in response to NDRG2 is functionally linked to NDRG2-induced cell differentiation. Previous study shows that Skp2 Ser72 (S72) phosphorylation is critical for Skp2 E3 ligase activity and function [26], we introduced WT Skp2 and a mutant Skp2 construct, which has a mutated S72 residue from serine to alanine (S72A), respectively. WT Skp2 readily decreased endogenous p21 and p27 expression levels, and further blocked the induction of p21 and p27 by NDRG2, whereas Skp2 S72A was profoundly impaired in this function (Fig. 7a). Thus, Skp2 E3 ligase activity is required for NDRG2-driven p21 and p27 upregulations. Moreover, Skp2 expression restored cell viability diminished by NDRG2, and blocked NDRG2-induced AKP activity following NaBT treatment in addition (Figs. 7b, c). In contrast, Skp2 S72A markedly decreased this activity. Furthermore, the reduction of endogenous Skp2 by small interfering RNA (siRNA) induced E-cadherin expression and AKP activity, and the effect was more prominent in the presence of NaBT (Figs. 7d, e). We conclude Skp2 functions as an E3 ligase involved in NDRG2 induction of CRC cell differentiation.
NDRG2 suppression of Skp2 controls cell differentiation. a-c We overexpressed wild-type Skp2 (wSkp2) and Skp2 S72A mutant (mSkp2) using relative expressing lentivirus in HT29 cells with or without NDRG2 expression. a Levels of the indicated proteins were determined in each cell population. b Cell viabilities in each group were determined by MTT assay. c AKP activity was determined in each cell population following a 48-h treatment of 2 mM NaBT. Data are expressed as means ±SD (n = 3). *P < 0.05. d, e HT29 cells were transfected with control or Skp2 siRNA. 24 h after transfection, cells were treated with 2 mM NaBT for additional 48 h. Indicated protein levels and AKP activity were determined relatively. Data are expressed as means ±SD (n = 3). *P < 0.05
Low NDRG2 expression and an NDRG2low/Skp2high gene expression signature correlate with poor patient outcome
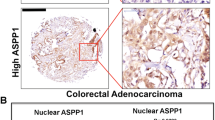
The clinical significance of the reciprocal relationship between NDRG2 and Skp2 was determined in 112 CRC patients. NDRG2 was broadly lowly expressed in CRCs compared with adjacent normal colon tissues, whereas the expression pattern of Skp2 was the inverse association of NDRG2 (Figs. 8a, b). Further, in silico analysis of 383 primary CRC tissues of the multidimensional data set from TCGA revealed a negative correlation of NDRG2 and Skp2 mRNA expression levels (Fig. 8c), confirming our propose that NDRG2 regulation of Skp2 in transcriptional level. Statistical analysis showed Skp2 was highly expressed in the more aggressive CRCs, in which NDRG2 was less stained (Figs. 8d, 1b). Moreover, Skp2 was predominantly highly expressed in poor differentiated CRCs, in which NDRG2 was diminished (Figs. 8a, e,1c). Therefore, the expression levels of NDRG2 and Skp2 were positively and negatively associated with CRC differentiation status respectively. That is to say, NDRG2 regulation of Skp2 plays key roles during CRC progression and differentiation.
Low NDRG2 expression and an NDRG2low/Skp2high gene expression signature correlate with poor patient outcome. a Immunohistochemistry staining of NDRG2 and Skp2 in human adjacent normal and cancerous colon tissues from CRC patients. b Negative correlation between NDRG2 and Skp2 expression levels with linear regression and Pearson’s correlation significance (P < 0.0001, ANOVA test). c Negative correlation between NDRG2 and Skp2 mRNA expression patterns in TCGA cbioportal data set with linear regression and Pearson’s correlation significance (P < 0.001, ANOVA test). d, e Skp2 expression by immunohistochemistry staining in human tissues at different stage d or differentiation status e. f, g Kaplan–Meier plots indicate the clinical outcomes for Skp2 expression f and NDRG2low/Skp2high expression signature g by immunohistochemistry staining in CRCs. n indicates the number of patient samples evaluated in each analysis. P-values were calculated using the Mantel–Cox log-rank test. h The proposed mechanism of NDRG2 action in regulation of CRC differentiation
It has been demonstrated that low NDRG2 expression correlated with decreased survival (Figs. 1e, f). Although Skp2 expression alone had no significant power in terms of overall survival (Fig. 8f), low NDRG2 expression showed a most evident correlation with decreased survival in patients whose tumors also had high Skp2 expression (Fig. 8g). We conclude that low NDRG2 expression correlates with poor prognosis; however, the negative effect of low NDRG2 expression is most evident when Skp2 levels are also high. Thus, the NDRG2low/Skp2high gene signature is correlated with poor clinical outcome in CRCs. The reciprocal regulatory relationship between NDRG2 and Skp2 contributes to CRC differentiation and aggressive phenotype.
Discussion
Differentiation has long been recognized to be of importance in solid tumors in which undifferentiated histology is usually a marker of tumor aggressiveness and poor prognosis. Confirming this point, we observed a high correlation between differentiation stage and survival in CRC patients (Fig. 1d). The association of NDRG2 with tumor differentiation has been recognized for long time [21, 22]. However, how NDRG2 confers to the differentiation process during tumor progression is largely unknown. Our study investigated the mechanism of NDRG2 coordinates colon epithelial cell differentiation and the role of its aberrant expression in CRC evolution. The data demonstrate for the first time that NDRG2 attenuates Skp2 expression through inhibiting β-catenin nuclear translocation, and consequently blocks the ubiquitinations and degradations of p21 and p27, which confers to the promotion of cell differentiation (Fig. 8h). Due to the high correlation of NDRG2low/Skp2high gene expression signature with poor CRC patient outcome, NDRG2-dependent repression of Skp2 presumably is the predominant route by which NDRG2 drives cell differentiation.
The diminished rate of cell cycle progression is one of the characters of differentiated cells. The Cip/Kip belongs to CDK inhibitor family, including p21/Cip1 and p27/Kip1. Accumulated evidence shows that elevated p21 or p27 expression causes growth inhibition in various cancers [27, 28]. Moreover, unlike p53, the frequency of cancer-related somatic mutations in the Cip/Kip genes is rare [29, 30], which underlines the importance of p21 and p27 expression regulation for cell transformation. We here observed that p21 and p27 are involved in NDRG2 effects on the blockage of cell growth and cell cycle, and the induction of differentiation in CRCs. The pivotal role of NDRG2 induction of p21 and p27 is supported by shRNA experiments where reducing p21 or p27 levels causes attenuation of NDRG2-induced cell differentiation and growth inhibition. The ablation of p21 had a greater effect on this reversal compared with p27, indicating that p21 might be the preferential functional target of NDRG2.
The increased protein stabilities of p21 and p27 by NDRG2 raise the possibility that this regulation is through E3 ligase Skp2, which was a critical regulator of cell cycle progression via targeting several CDK inhibitors for degradation. We first observed a high correlation between NDRG2 induction and Skp2 reduction in either long-term or short-term induced CRC cell differentiation models. Further, NDRG2 is shown to repress Skp2 both on mRNA and protein levels, indicating NDRG2 regulating Skp2 transcription. The involvement of Skp2 in NDRG2-induced differentiation was confirmed with rescue expression of WT Skp2 and mutant Skp2 S72A in CRC cells. S72A mutation can evade Akt-mediated phosphorylation of Skp2, which was crucial for the SCF complex formation and the E3 ligase activity [26]. As expected, WT Skp2 reversed NDRG2 induction of CRC differentiation and growth inhibition, whereas Skp2 S72A cannot. Contrary to p27, which is regulated through protein stabilization, p21 is found regulated at both the transcriptional and post-transcriptional levels by NDRG2 expression. However, the increased mRNA levels of p21 are not sufficient in itself to explain the strong induction of its protein levels by NDRG2. Thus, NDRG2 repression of Skp2 is still the primary route for increased p21 and p27 expression levels. Beyond that, Skp2 has also been shown regulate a number of biological behaviors, including epithelial–mesenchymal transition, glycolysis, and cancer stem cell traits [15, 31], which indicates the comprehensive role of NDRG2 in regulation of Skp2 besides differentiation.
It is widely accepted that WNT pathway plays a central role in the etiology of CRCs [2, 4, 24]. The activation of WNT signal induces nuclear translocation of β-catenin through the repression of APC/axin/GSK-3β complex, and further activates oncogenic signals by β-catenin/TCF complexes. Skp2 is one of the important targets that is transcriptionally regulated by WNT pathway. The activated β-catenin/TCF complexes induce Skp2 transcription via the recognition of two TCF/LEF1 consensus binding sites in Skp2 promoter [16]. As NDRG2 has been shown represses β-catenin and TCF/LEF activities in previous study [32], it is reasonable to assume that NDRG2 suppression of Skp2 transcription is through the blockage of WNT/β-catenin pathway. Confirming this point, NDRG2 expression dramatically reduced the total and nucleus fraction of β-catenin. Accordingly, the occupancies of TCF/LEF complex on Skp2 promoter were markedly decreased in the presence of NDRG2. Further, with the introduction of GSK-3β inhibitor, we found NDRG2 suppression of Skp2 was dependent on GSK-3β activation. It is reported that NDRG2 decreased PI3K/AKT activity through the regulation of PTEN phosphatase activity, and activated GSK-3β in turn [25]. Thus, NDRG2-dependent activation of GSK-3β through PI3K/AKT pathway was at least one of the routes in terms of suppression of Skp2.
We and other groups found highly NDRG2 expression (NDRG2high) were correlated with better CRC outcome [33, 34]. In this study, we further confirmed this result both in a cohort study of CRC tissues from 112 patients and TCGA data set. Thus, decreased NDRG2 levels appeared to be a relatively early event in tumorigenesis and low NDRG2 levels correlated with worse prognosis later in tumorigenesis. Skp2 overexpression was a feature of poor differentiated tumors [35, 36], and a previous study showed that Skp2 correlated with worse clinical outcome [37]. However, although there was a tendency of better survival in reduced SKP2 expression of CRC patients, we did not confirm high Skp2 levels correlated with overall survival. Noticeably, we found that an NDRG2low/Skp2high gene signature was highly correlated with poor clinical outcome in CRCs. Although low NDRG2 expression correlates with poor prognosis, the negative effect of reduced NDRG2 expression was most evident with increased Skp2 levels. These results validated the pathological significance of the reciprocal regulatory relationship between NDRG2 and Skp2 described here and suggest that the deleterious effects of low NDRG2 expression are exacerbated by high Skp2 expression. Further, we also observed a highly correlation of NDRG2high/p21high or NDRG2high/p27high gene signature with a better outcome from CRCs in TCGA data set, which verified the clinical significance of Skp2-p21/p27 axis in NDRG2-induced CRC differentiation.
It is widely accepted that NDRG2 contains a putative α/β-hydrolase fold domain (NDR domain), but lacks the expected enzymatic catalytic motif [38]. Thus, the key region of NDRG2 function as a tumor suppressor is not limited to homeodomain of α/β-superfamily. We here generated NDRG2 deletion mutants without either NH2-terminal half or carboxyl-terminal half. The loss of NH2-terminal domain can completely abolish NDRG2 repression of Skp2 and induction of AKP activity, indicating the necessary of NH2-terminal domain for NDRG2-induced CRC differentiation. Further studies are needed to fully address the specific functional motifs in the NH2-terminal half of NDRG2.
Taken together, out data provide the first evidence that NDRG2 functions as a critical regulator of CRC differentiation. NDRG2 suppresses β-catenin nuclear translocation, and blocks β-catenin/TCF/LEF complex transcriptional activity on Skp2 promoter, which in turn increases p21 and p27 stabilities and expression levels via Skp2 suppression. As such, we point out an NDRG2low/Skp2high gene expression signature is correlated with poor patient outcome and could be considered as a diagnostic marker of CRCs.
Materials and methods
Tissue samples and immunohistochemistry staining
This study was approved by the ethics committee of the Fourth Military Medical University. All 112 patients and the 681 subjects in the CRC sample study cohort provided full consent for the study. The tumor diagnosis and immunohistochemistry staining were performed as described previously [20].
Animals
Ndrg2 conditional knockout (Ndrg2-/-) were generated by Shanghai Biomodel Organism Science & Technology Development Co., Ltd and maintained on C57BL6/J background. All animals were raised under specific pathogen-free conditions. One- to 3-month-old mice were used in experiments. WT mice and Ndrg2-/- mice were age-matched and all mice used were male. Typically, six mice were used for each group of treatment and all experiments were repeated at least three times.
Immunofluorescence assay
For immunofluorescent staining, the whole colon was taken and fixed with formalin, incubated with indicated antibodies, and following fluorescein isothiocyanate (FITC)-conjugated secondary antibodies and counterstained with 4',6-diamidino-2-phenylindole (DAPI). The sections were captured by confocal laser scanning microscopy (Nikon, Japan) and the fluorescence intensity of immunofluorescence images were measured by ImageJ for quantification of five areas per slide.
Plasmid construction
DNA fragments encoding human NDRG2 deletion mutant without NH2-terminal half (ΔN) or carboxyl-terminal half (ΔC) were amplified by PCR from the pcDNA3.1-NDRG2 plasmid. WT pBabe-Skp2 and pBabe-Skp2 S72A mutant were kindly gifts from Dr. Hui-kuan Lin (The University of Texas MD Anderson Cancer Center, Houston, USA). S72 is the site for Akt-mediated Skp2 activation in vivo. Previous study confirmed that mutation of S72 site is critical to abolish Skp2-mediated target protein degradation. SKP2 and control siRNA are 21-bp synthetic molecules corresponding to SKP2 and the firefly luciferase gene relatively. The p21 shRNA sequence used to knockdown p21 expression was 5′-CCGGGAGCGATGGAACTTCGACTTTCTCGAGAAAGTCGAAGTTCCATCGCTCTTTTTG-3′ (p21-sh233) and 5’-CCGGGACAGATTTCTACCACTCCAACTCGAGTTGGAGTGGTAGAAATCTGTCTTTTTG-3′ (p21-sh535). The p27 shRNA sequence used to knockdown p27 expression was 5′-CCGGGAGCAATGCGCAGGAATAAGGCTCGAGCCTTATTCCTGCGCATTGCTCTTTTTG-3′. (Shanghai GenePharma). Scramble was used as negative control. NDRG2-overexpressing and shRNA lentivirus were generated and infected as described previously [39].
Luciferase reporter assay
HT29 cells were transfected with TOPflash or FOPflash and the indicated protein-expressing vectors. In all, 20 mM of LiCl (Sigma) was added to the medium for 12 h. Then cells were lysed according to the manufacturer’s instructions of dual-luciferase reagent assay kit (Promega, Madison, WI).
Cell culture and differentiation induction and analysis
HT29, HCT116, and Caco-2 cells were purchased from ATCC and cultured in Dulbecco’s modified Eagle's medium (DMEM) supplemented with 10% of fetal calf serum. Cell differentiation was induced as described previously [22]. For the intestinal AKP activity assay, the activity was measured by IAP kit (Jiancheng-Nanjing, China) according to the manual instructions.
Cell growth and viability assays
MTT assay, EDU incorporation assay, and cell cycle assay were performed as described previously [19, 20, 22]. For serum starvation, cells were incubated in DMEM without serum for 72 h, and then the medium was replaced with 10% fetal bovine serum medium for the times indicated.
Quantitative PCR
Extract the RNA and generate complementary DNA through GoScript Reverse Transcription System (Promega). The quantitative PCR assay was performed as described previously [40]. Primer sequences are available on request.
In vivo ubiquitination assay
Control or NDRG2-overexpressing HT29 cells were transfected with pcDNA3.1, pcDNA3.1-p21, or pcDNA3.1-p27 accompanied with pcDNA3.1(+)−3 × HA-Ub. The in vivo ubiquitination assay was then performed as described previously [41].
Western blotting
The cells were lysed and the concentrations were measured by BCA™ protein assay kit (Thermo Scientific). In all, 40 μg proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to nitrocellulose membranes (Hybond ECL). Primary antibodies were then introduced at dilutions of 1:1000 for anti-Skp2 (Santa Cruz Biotechnology, sc-7164), 1:1000 for anti-NDRG2 (ABNOVA, H00057447-M03), 1:1000 for anti-p21 (Cell Signaling Technology, 2947), 1:1000 for anti-p27 (Cell Signaling Technology, 2552), 1:500 for anti-E-cadherin (BD Bioscience, MAB1838), anti-Cyclin D1 (Cell Signaling Technology, 2922), anti-CDK2 (Abcam, ab32147), anti-CDK4 (Abcam, ab108357), anti-p-Akt (Ser473) (Cell Signaling Technology, 4060), anti-Akt (Cell Signaling Technology, 2938), anti-p-GSK-3β(Ser9) (Cell Signaling Technology, 9322), anti-GSK-3β (Cell Signaling Technology, 9315), anti-β-catenin (Sigma, C2206), and anti-β-actin (Sigma, A5441). Secondary antibodies were then added against the primary antibodies. The blots were detected by chemiluminescence (Pierce) or Odyssey Imaging System (Li-Cor Biosciences, NE).
Chromatin immunoprecipitation
The chromatin immunoprecipitation (ChIP) analysis was performed using the ChIP Assay kit (Upstate Biotechnology, Charlottesville, VA, USA) as described previously [20]. The sequences of PCR primers detecting Skp2 promoter are available on request.
Protein degradation assay
NDRG2-overexpressing HT29 and Caco-2 cells and control cells were incubated with CHX (10 μg/ml, Sigma) for indicated time. Cells were then harvest and western blotting was performed as described above.
In vivo tumorigenicity assay
Four- to 6-week-old athymic mice (four mice per group) were injected subcutaneously indicated cells (1 × 107 cells per mouse). Tumor volume was measured and calculated as indicated [41].
TCGA patient data analysis
The Cancer Genome Atlas cBioPortal was used to determine overall patient survival and the tendency for co-occurrence of NDRG2, Skp2, p21, and p27 alterations in the data set for Colorectal Adenocarcinoma (TCGA, Provisional). NDRG2, Skp2, p21, and p27 were entered as the query genes and overall survival and co-expression was provided through the cBioPortal user interface. The raw data were downloaded from TCGA through the cBioPortal and manually graphed in Prism (Graphpad) with relative mRNA alterations. For gene expression association analysis, gene expression levels were normalized for each patient to the median expression across all patients. A log conversion was applied to the normalized, averaged NDRG2 mRNA and Skp2 mRNA expression levels and then plotted as a correlation graph in Prism software with a linear regression curve.
Survival analysis
Survival analysis was determined with CRC patient progression-free survival data from FMMU or TCGA data set. Patient data with valid gene mRNA or protein expression levels were used to estimate medians and bounds for upper and lower means. Kaplan–Meier survival graphs were plotted, and log-rank tests were performed.
Statistical analysis
Data are expressed as mean±SD. Statistical analysis was performed with the SPSS10.0 software package by using Student’s t-test for independent groups. Statistical significance was based on a value of P ≤ 0.05.
References
Derwinger K, Kodeda K, Bexe-Lindskog E, Taflin H. Tumour differentiation grade is associated with TNM staging and the risk of node metastasis in colorectal cancer. Acta Oncol (Madr). 2010;49:57–62.
Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–20.
Goss KH, Groden J. Biology of the adenomatous polyposis coli tumor suppressor. J Clin Oncol. 2000;18:1967–79.
Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–90.
Ng RC, Matsumaru D, Ho AS, Garcia-Barcelo MM, Yuan ZW, Smith D, et al. Dysregulation of Wnt inhibitory factor 1 (Wif1) expression resulted in aberrant Wnt-beta-catenin signaling and cell death of the cloaca endoderm, and anorectal malformations. Cell Death Differ. 2014;21:978–89.
Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–804.
Marikawa Y, Elinson RP. Beta-TrCP is a negative regulator of Wnt/beta-catenin signaling pathway and dorsal axis formation in Xenopus embryos. Mech Dev. 1998;77:75–80.
He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12.
Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–6.
Pfeuty B, David-Pfeuty T, Kaneko K. Underlying principles of cell fate determination during G1 phase of the mammalian cell cycle. Cell Cycle. 2008;7:3246–57.
Steinman RA, Hoffman B, Iro A, Guillouf C, Liebermann DA, el-Houseini ME. Induction ofp21 (WAF-1/CIP1) during differentiation. Oncogene. 1994;9:3389–96.
Kim SJ, Lee HW, Gu Kang H, La SH, Choi IJ, Ro JY, et al. Ablation of galectin-3 inducesp27(KIP1)-dependent premature senescence without oncogenic stress. Cell Death Differ. 2014;21:1769–79.
Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol. 2005;16:323–33.
Gao D, Inuzuka H, Tseng A, Chin RY, Toker A, Wei W. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat Cell Biol. 2009;11:397–408.
Qu X, Shen L, Zheng Y, Cui Y, Feng Z, Liu F, et al. A signal transduction pathway from TGF-beta1 to SKP2 via Akt1 and c-Myc and its correlation with progression in human melanoma. J Invest Dermatol. 2014;134:159–67.
Tang Y, Simoneau AR, Liao WX, Yi G, Hope C, Liu F, et al. WIF1, a Wnt pathway inhibitor, regulates SKP2 and c-myc expression leading to G1 arrest and growth inhibition of human invasive urinary bladder cancer cells. Mol Cancer Ther. 2009;8:458–68.
Deng Y, Yao L, Chau L, Ng SS, Peng Y, Liu X, et al. N-Myc downstream-regulated gene 2 (NDRG2) inhibits glioblastoma cell proliferation. Int J Cancer. 2003;106:342–7.
Lorentzen A, Vogel LK, Lewinsky RH, Saebo M, Skjelbred CF, Godiksen S, et al. Expression of NDRG2 is down-regulated in high-risk adenomas and colorectal carcinoma. BMC Cancer. 2007;7:192.
Xu X, Li J, Sun X, Guo Y, Chu D, Wei L, et al. Tumor suppressor NDRG2 inhibits glycolysis and glutaminolysis in colorectal cancer cells by repressing c-Myc expression. Oncotarget. 2015;6:26161–76.
Shen L, Qu X, Ma Y, Zheng J, Chu D, Liu B, et al. Tumor suppressor NDRG2 tips the balance of oncogenic TGF-beta via EMT inhibition in colorectal cancer. Oncogenesis. 2014;3:e86.
Kang K, Nam S, Kim B, Lim JH, Yang Y, Lee MS, et al. Inhibition of osteoclast differentiation by overexpression of NDRG2 in monocytes. Biochem Biophys Res Commun. 2015;468:611–6.
Zhang J, Li F, Liu X, Shen L, Liu J, Su J, et al. The repression of human differentiation-related gene NDRG2 expression by Myc via Miz-1-dependent interaction with the NDRG2 core promoter. J Biol Chem. 2006;281:39159–68.
Evers BM, Ko TC, Li J, Thompson EA. Cell cycle protein suppression and p21 induction in differentiating Caco-2 cells. Am J Physiol. 1996;271:G722–727.
Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–7.
Nakahata S, Ichikawa T, Maneesaay P, Saito Y, Nagai K, Tamura T, et al. Loss of NDRG2 expression activates PI3K-AKT signalling via PTEN phosphorylation in ATLL and other cancers. Nat Commun. 2014;5:3393.
Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, et al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat Cell Biol. 2009;11:420–32.
Chen J, Willingham T, Shuford M, Bruce D, Rushing E, Smith Y, et al. Effects of ectopic overexpression ofp21(WAF1/CIP1) on aneuploidy and the malignant phenotype of human brain tumor cells. Oncogene. 1996;13:1395–403.
Craig C, Wersto R, Kim M, Ohri E, Li Z, Katayose D, et al. A recombinant adenovirus expressing p27Kip1 induces cell cycle arrest and loss of cyclin-Cdk activity in human breast cancer cells. Oncogene. 1997;14:2283–9.
Ponce-Castaneda MV, Lee MH, Latres E, Polyak K, Lacombe L, Montgomery K, et al. p27Kip1: chromosomal mapping to 12p12-12p13.1 and absence of mutations in human tumors. Cancer Res. 1995;55:1211–4.
Shiohara M, Koike K, Komiyama A, Koeffler HP. p21WAF1 mutations and human malignancies. Leuk Lymphoma. 1997;26:35–41.
Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556–68.
Kim YJ, Yoon SY, Kim JT, Song EY, Lee HG, Son HJ, et al. NDRG2 expression decreases with tumor stages and regulates TCF/beta-catenin signaling in human colon carcinoma. Carcinogenesis. 2009;30:598–605.
Chu D, Zhang Z, Li Y, Wu L, Zhang J, Wang W, et al. Prediction of colorectal cancer relapse and prognosis by tissue mRNA levels of NDRG2. Mol Cancer Ther. 2011;10:47–56.
Kim YJ, Kang HB, Yim HS, Kim JH, Kim JW. NDRG2 positively regulates E-cadherin expression and prolongs overall survival in colon cancer patients. Oncol Rep. 2013;30:1890–8.
Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer. 2008;112:1415–24.
Shapira M, Ben-Izhak O, Linn S, Futerman B, Minkov I, Hershko DD. The prognostic impact of the ubiquitin ligase subunits Skp2 and Cks1 in colorectal carcinoma. Cancer. 2005;103:1336–46.
Tian YF, Chen TJ, Lin CY, Chen LT, Lin LC, Hsing CH, et al. SKP2 overexpression is associated with a poor prognosis of rectal cancer treated with chemoradiotherapy and represents a therapeutic target with high potential. Tumour Biol. 2013;34:1107–17.
Yao L, Zhang J, Liu X. NDRG2: a Myc-repressed gene involved in cancer and cell stress. Acta Biochim Biophys Sin (Shanghai). 2008;40:625–35.
Guo Y, Ma J, Wu L, Wang Q, Li X, Li X, et al. Hyperthermia-induced NDRG2 upregulation inhibits the invasion of human hepatocellular carcinoma via suppressing ERK1/2 signaling pathway. PLoS ONE. 2013;8:e61079
Shen L, O’Shea JM, Kaadige MR, Cunha S, Wilde BR, Cohen AL, et al. Metabolic reprogramming in triple-negative breast cancer through Myc suppression of TXNIP. Proc Natl Acad Sci USA. 2015;112:5425–30.
Li X, Shen L, Zhang J, Su J, Shen L, Liu X, et al. Degradation of HER2 by Cbl-based chimeric ubiquitin ligases. Cancer Res. 2007;67:8716–24.
Acknowledgements
This work was financially sponsored by grants from the National Natural Science Foundation of China (no. 31401161; no. 81230043; no. 31571437; no. 81502370; no. 81401069; no. 81770523; and no. 81502210), the Founds for Creative Research Groups of China (no. 81421003), and the State Key Laboratory of Cancer Biology Project (CBSKL2017Z11).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. If you remix, transform, or build upon this article or a part thereof, you must distribute your contributions under the same license as the original. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/.
About this article
Cite this article
Shen, L., Qu, X., Li, H. et al. NDRG2 facilitates colorectal cancer differentiation through the regulation of Skp2-p21/p27 axis. Oncogene 37, 1759–1774 (2018). https://doi.org/10.1038/s41388-017-0118-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-017-0118-7
This article is cited by
-
The involvement of E3 ubiquitin ligases in the development and progression of colorectal cancer
Cell Death Discovery (2023)
-
Intact regulation of G1/S transition renders esophageal squamous cell carcinoma sensitive to PI3Kα inhibitors
Signal Transduction and Targeted Therapy (2023)
-
MESP2 binds competitively to TCF4 to suppress gastric cancer progression by regulating the SKP2/p27 axis
Cell Death Discovery (2023)
-
BCR-ABL triggers a glucose-dependent survival program during leukemogenesis through the suppression of TXNIP
Cell Death & Disease (2023)
-
MYOD-SKP2 axis boosts tumorigenesis in fusion negative rhabdomyosarcoma by preventing differentiation through p57Kip2 targeting
Nature Communications (2023)