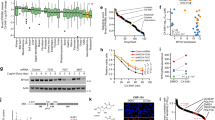
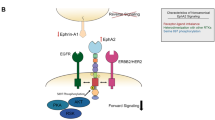
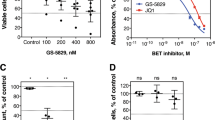
Abstract
Ewing sarcoma (ES) is an aggressive bone and soft tissue malignancy that predominantly affects children and adolescents. CD99 is a cell surface protein that is highly expressed on ES cells and is required to maintain their malignancy. We screened small molecule libraries for binding to extracellular domain of recombinant CD99 and subsequent inhibition of ES cell growth. We identified two structurally similar FDA-approved compounds, clofarabine and cladribine that selectively inhibited the growth of ES cells in a panel of 14 ES vs. 28 non-ES cell lines. Both drugs inhibited CD99 dimerization and its interaction with downstream signaling components. A membrane-impermeable analog of clofarabine showed similar cytotoxicity in culture, suggesting that it can function through inhibiting CD99 independent of DNA metabolism. Both drugs drastically inhibited anchorage-independent growth of ES cells, but clofarabine was more effective in inhibiting growth of three different ES xenografts. Our findings provide a novel molecular mechanism for clofarabine that involves direct binding to a cell surface receptor CD99 and inhibiting its biological activities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kovar H. Ewing’s sarcoma and peripheral primitive neuroectodermal tumors after their genetic union. Curr Opin Oncol. 1998;10:334–42.
Arndt CA, Crist WM. Common musculoskeletal tumors of childhood and adolescence. N Engl J Med. 1999;341:342–52.
Kovar H, Amatruda J, Brunet E, Burdach S, Cidre-Aranaz F, de Alava E, et al. The second European interdisciplinary Ewing sarcoma research summit—a joint effort to deconstructing the multiple layers of a complex disease. Oncotarget. 2016;7:8613–24.
Lawlor ER, Sorensen PH. Twenty years on: What do we really know about ewing sarcoma and what is the path forward? Crit Rev Oncog. 2015;20:155–71.
Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Le Deley MC, et al. Ewing sarcoma: Current management and future approaches through collaboration. J Clin Oncol. 2015;33:3036–46.
Longhi A, Ferrari S, Tamburini A, Luksch R, Fagioli F, Bacci G, et al. Late effects of chemotherapy and radiotherapy in osteosarcoma and Ewing sarcoma patients: the Italian Sarcoma Group Experience (1983–2006). Cancer. 2012;118:5050–9.
Kovar H, Dworzak M, Strehl S, Schnell E, Ambros IM, Ambros PF, et al. Overexpression of the pseudoautosomal gene MIC2 in Ewing’s sarcoma and peripheral primitive neuroectodermal tumor. Oncogene. 1990;5:1067–70.
Ambros IM, Ambros PF, Strehl S, Kovar H, Gadner H, Salzer-Kuntschik M. MIC2 is a specific marker for Ewing’s sarcoma and peripheral primitive neuroectodermal tumors. Evidence for a common histogenesis of Ewing’s sarcoma and peripheral primitive neuroectodermal tumors from MIC2 expression and specific chromosome aberration. Cancer. 1991;67:1886–93.
Fellinger EJ, Garin-Chesa P, Triche TJ, Huvos AG, Rettig WJ. Immunohistochemical analysis of Ewing’s sarcoma cell surface antigen p30/32MIC2. Am J Pathol. 1991;139:317–25.
Perlman EJ, Dickman PS, Askin FB, Grier HE, Miser JS, Link MP. Ewing’s sarcoma—routine diagnostic utilization of MIC2 analysis: a Pediatric Oncology Group/Children’s Cancer Group Intergroup Study. Hum Pathol. 1994;25:304–7.
Weidner N, Tjoe J. Immunohistochemical profile of monoclonal antibody O13: antibody that recognizes glycoprotein p30/32MIC2 and is useful in diagnosing Ewing’s sarcoma and peripheral neuroepithelioma. Am J Surg Pathol. 1994;18:486–94.
Lee CS, Southey MC, Waters K, Kannourakis G, Georgiou T, Armes JE, et al. EWS/FLI-1 fusion transcript detection and MIC2 immunohistochemical staining in the diagnosis of Ewing’s sarcoma. Pediatr Pathol Lab Med 1996;16:379–92.
Halliday BE, Slagel DD, Elsheikh TE, Silverman JF. Diagnostic utility of MIC-2 immunocytochemical staining in the differential diagnosis of small blue cell tumors. Diagn Cytopathol. 1998;19:410–6.
Lucas DR, Bentley G, Dan ME, Tabaczka P, Poulik JM, Mott MP. Ewing sarcoma vs lymphoblastic lymphoma. A comparative immunohistochemical study. Am J Clin Pathol. 2001;115:11–17.
Khoury JD. Ewing sarcoma family of tumors. Adv Anat Pathol. 2005;12:212–20.
Aubrit F, Gelin C, Pham D, Raynal B, Bernard A. The biochemical characterization of E2, a T cell surface molecule involved in rosettes. Eur J Immunol. 1989;19:1431–6.
Gelin C, Aubrit F, Phalipon A, Raynal B, Cole S, Kaczorek M, et al. The E2 antigen, a 32 kd glycoprotein involved in T-cell adhesion processes, is the MIC2 gene product. EMBO J. 1989;8:3253–9.
Ellis NA, Tippett P, Petty A, Reid M, Weller PA, Ye TZ, et al. PBDX is the XG blood group gene. Nat Genet. 1994;8:285–90.
Suh YH, Shin YK, Kook MC, Oh KI, Park WS, Kim SH, et al. Cloning, genomic organization, alternative transcripts and expression analysis of CD99L2, a novel paralog of human CD99, and identification of evolutionary conserved motifs. Gene. 2003;307:63–76.
Bixel G, Kloep S, Butz S, Petri B, Engelhardt B, Vestweber D. Mouse CD99 participates in T-cell recruitment into inflamed skin. Blood. 2004;104:3205–13.
Dworzak MN, Fritsch G, Fleischer C, Printz D, Froschl G, Buchinger P, et al. CD99 (MIC2) expression in paediatric B-lineage leukaemia/lymphoma reflects maturation-associated patterns of normal B-lymphopoiesis. Br J Haematol. 1999;105:690–5.
Schenkel AR, Mamdouh Z, Chen X, Liebman RM, Muller WA. CD99 plays a major role in the migration of monocytes through endothelial junctions. Nat Immunol. 2002;3:143–50.
Hu-Lieskovan S, Zhang J, Wu L, Shimada H, Schofield DE, Triche TJ. EWS-FLI1 fusion protein up-regulates critical genes in neural crest development and is responsible for the observed phenotype of Ewing’s family of tumors. Cancer Res. 2005;65:4633–44.
Miyagawa Y, Okita H, Nakaijima H, Horiuchi Y, Sato B, Taguchi T, et al. Inducible expression of chimeric EWS/ETS proteins confers Ewing’s family tumor-like phenotypes to human mesenchymal progenitor cells. Mol Cell Biol. 2008;28:2125–37.
Rocchi A, Manara MC, Sciandra M, Zambelli D, Nardi F, Nicoletti G, et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. J Clin Invest. 2010;120:668–80.
Franzetti GA, Laud-Duval K, Bellanger D, Stern MH, Sastre-Garau X, Delattre O. MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor. Oncogene. 2013;32:3915–21.
Ventura S, Aryee DN, Felicetti F, De Feo A, Mancarella C, Manara MC, et al. CD99 regulates neural differentiation of Ewing sarcoma cells through miR-34a-Notch-mediated control of NF-kappaB signaling. Oncogene. 2016;35:3944–54.
Sohn HW, Choi EY, Kim SH, Lee IS, Chung DH, Sung UA, et al. Engagement of CD99 induces apoptosis through a calcineurin-independent pathway in Ewing’s sarcoma cells. Am J Pathol. 1998;153:1937–45.
Scotlandi K, Baldini N, Cerisano V, Manara MC, Benini S, Serra M, et al. CD99 engagement: an effective therapeutic strategy for Ewing tumors. Cancer Res. 2000;60:5134–42.
Kreppel M, Aryee DN, Schaefer KL, Amann G, Kofler R, Poremba C, et al. Suppression of KCMF1 by constitutive high CD99 expression is involved in the migratory ability of Ewing’s sarcoma cells. Oncogene. 2006;25:2795–2800.
Scotlandi K, Perdichizzi S, Bernard G, Nicoletti G, Nanni P, Lollini PL, et al. Targeting CD99 in association with doxorubicin: an effective combined treatment for Ewing’s sarcoma. Eur J Cancer. 2006;42:91–6.
Manara MC, Bernard G, Lollini PL, Nanni P, Zuntini M, Landuzzi L, et al CD99 acts as an oncosuppressor in osteosarcoma. Mol Biol Cell. 2006;17:1910–21.
Zucchini C, Manara MC, Pinca RS, De Sanctis P, Guerzoni C, Sciandra M, et al. CD99 suppresses osteosarcoma cell migration through inhibition of ROCK2 activity. Oncogene. 2014;33:1912–21.
Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54.
Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;10:e1004475.
Choi G, Lee SW, Jung KC, Choi EY. Detection of homodimer formation of CD99 through extracelluar domain using bimolecular fluorescence complementation analysis. Exp Mol Med. 2007;39:746–55.
Lee MK, Kim HS, Kim SS, Cho MH, Lee IS. Analysis of the dimerization of human CD99 using bimolecular fluorescence complementation technique. J Microbiol Biotechnol. 2008;18:472–6.
Kim HJ, Chong KH, Kang SW, Lee JR, Kim JY, Hahn MJ, et al. Identification of cyclophilin A as a CD99-binding protein by yeast two-hybrid screening. Immunol Lett. 2004;95:155–9.
Watson RL, Buck J, Levin LR, Winger RC, Wang J, Arase H, et al. Endothelial CD99 signals through soluble adenylyl cyclase and PKA to regulate leukocyte transendothelial migration. J Exp Med. 2015;212:1021–41.
Gollnest T, de Oliveira TD, Schols D, Balzarini J, Meier C. Lipophilic prodrugs of nucleoside triphosphates as biochemical probes and potential antivirals. Nat Commun. 2015;6:8716.
Gollnest T, Dinis de Oliveira T, Rath A, Hauber I, Schols D, Balzarini J, et al. Membrane-permeable triphosphate prodrugs of nucleoside analogues. Angew Chem. 2016;55:5255–8.
Gellini M, Ascione A, Flego M, Mallano A, Dupuis ML, Zamboni S, et al. Generation of human single-chain antibody to the CD99 cell surface determinant specifically recognizing Ewing’s sarcoma tumor cells. Curr Pharm Biotechnol. 2013;14:449–63.
Guerzoni C, Fiori V, Terracciano M, Manara MC, Moricoli D, Pasello M, et al. CD99 triggering in Ewing sarcoma delivers a lethal signal through p53 pathway reactivation and cooperates with doxorubicin. Clin Cancer Res. 2015;21:146–56.
Moricoli D, Carbonella DC, Dominici S, Fiori V, Balducci MC, Guerzoni C, et al. Process development of a human recombinant diabody expressed in E. coli: engagement of CD99-induced apoptosis for target therapy in Ewing’s sarcoma. Appl Microbiol Biotechnol. 2016;100:3949–63.
Bonate PL, Arthaud L, Cantrell WR Jr., Stephenson K, Secrist JA 3rd, Weitman S. Discovery and development of clofarabine: a nucleoside analogue for treating cancer. Nat Rev Drug Discov. 2006;5:855–63.
Sigal DS, Miller HJ, Schram ED, Saven A. Beyond hairy cell: the activity of cladribine in other hematologic malignancies. Blood. 2010;116:2884–96.
Kawasaki H, Carrera CJ, Piro LD, Saven A, Kipps TJ, Carson DA. Relationship of deoxycytidine kinase and cytoplasmic 5’-nucleotidase to the chemotherapeutic efficacy of 2-chlorodeoxyadenosine. Blood. 1993;81:597–601.
Chung SS, Eng WS, Hu W, Khalaj M, Garrett-Bakelman FE, Tavakkoli M, et al. CD99 is a therapeutic target on disease stem cells in myeloid malignancies. Sci Transl Med. 2017;9:eaaj2025.
Bernard A, Aubrit F, Raynal B, Pham D, Boumsell LA. T cell surface molecule different from CD2 is involved in spontaneous rosette formation with erythrocytes. J Immunol. 1988;140:1802–7.
Dworzak MN, Fritsch G, Buchinger P, Fleischer C, Printz D, Zellner A, et al. Flow cytometric assessment of human MIC2 expression in bone marrow, thymus, and peripheral blood. Blood. 1994;83:415–25.
Husak Z, Printz D, Schumich A, Potschger U, Dworzak MN. Death induction by CD99 ligation in TEL/AML1-positive acute lymphoblastic leukemia and normal B cell precursors. J Leukoc Biol. 2010;88:405–12.
Husak Z, Dworzak MN. CD99 ligation upregulates HSP70 on acute lymphoblastic leukemia cells and concomitantly increases NK cytotoxicity. Cell death Dis. 2012;3:e425.
Cox CV, Diamanti P, Moppett JP, Blair A. Investigating CD99 expression in leukemia propagating cells in childhood T cell acute lymphoblastic leukemia. PLoS One. 2016;11:e0165210.
Chung SS, Tavakkoli M, Devlin SM, Park CY. CD99 Is a therapeutic target on disease stem cells in acute myeloid leukemia and the myelodysplastic syndromes. Blood. 2013;122:2891.
Chung SS, Tavakkoli M, Klimek VM, Park CY. CD99 is a therapeutic target on disease initiating stem cells in acute myeloid leukemia and the myelodysplastic syndromes. Blood. 2014;124:3760.
Winger RC, Harp CT, Chiang MY, Sullivan DP, Watson RL, Weber EW, et al. Cutting edge: CD99 is a novel therapeutic target for control of T cell-mediated central nervous system autoimmune disease. J Immunol. 2016;196:1443–8.
Giovannoni G, Comi G, Cook S, Rammohan K, Rieckmann P, Soelberg Sorensen P, et al. A placebo-controlled trial of oral cladribine for relapsing multiple sclerosis. N Engl J Med. 2010;362:416–26.
Celik H, Sajwan KP, Selvanathan SP, Marsh BJ, Pai AV, Kont YS, et al. Ezrin binds to DEAD-box RNA helicase DDX3 and regulates its function and protein level. Mol Cell Biol. 2015;35:3145–62.
Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5.
Acknowledgements
We thank the NCI/DPT Open Chemical Repository for providing compounds. We thank Dr. Abraham T. Kallarakal for help with the SPR screening experiments, Kelli Schanze for assistance with the animal experiments and Giulia Ricci for technical assistance. We thank Dr. Geeta Upadhyay for providing us the purified Ly6k protein. The authors thank Dr. Chand Khanna from the NCI/NIH (Bethesda, MD) for OS cell lines (HOS-MNNG and MG63.3), Dr. Eugenie S. Kleinerman from the University of Texas MD Anderson Cancer Center (Houston, TX) for human OS cell lines (SAOS-2 and SAOS-2/LM7), Dr. David M. Loeb from the Johns Hopkins university (Baltimore, MD) for MHH-ES cells, Dr. Heinrich Kovar from the Children’s Cancer Research Institute, St. Anna Kinderkrebsforschung (Vienna, Austria) for STA-ET-7.2 cells, Dr. Timothy J. Triche from the Children’s Hospital (Los Angeles, CA) for 6647 cells, Dr. Angelo Rosolen from the University of Padova (Italy) and Dr. David N. Shapiro from the St. Jude Children’s Hospital (Memphis, TN) for rhabdomyosarcoma cell lines RH1, RH30 and RH4.
Funding
This work was supported by the funds from the Children's Cancer Foundation (to A. Üren), Hyundai Hope on Wheels (to A. Üren), the Alan B. Slifka Foundation (to A. Üren and K. Scotlandi) and the Italian Association for Cancer Research (AIRC_IG14049 to K. Scotlandi). The authors thank the Animal Models Shared Resource, Tissue Culture Shared Resource, Histopathology and Tissue Shared Resource and the Biacore Molecular Interaction Shared Resource at the Lombardi Comprehensive Cancer Center (Georgetown University), which are supported by a grant P30CA51008 from the National Cancer Institute.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Georgetown University has filed a patent application for the use of CD99 inhibitors in the treatment of Ewing sarcoma, in which Drs. AÜ, HÇ, and JAT was listed as inventors.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Çelik, H., Sciandra, M., Flashner, B. et al. Clofarabine inhibits Ewing sarcoma growth through a novel molecular mechanism involving direct binding to CD99. Oncogene 37, 2181–2196 (2018). https://doi.org/10.1038/s41388-017-0080-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-017-0080-4
This article is cited by
-
Current Challenges and Potential Strategies for Designing a New Generation of Chimeric Antigen Receptor-T cells with High Anti-tumor Activity in Solid Tumors
Current Tissue Microenvironment Reports (2023)
-
SPRD: a surface plasmon resonance database of common factors for better experimental planning
BMC Molecular and Cell Biology (2021)
-
Gene expression profile association with poor prognosis in epithelial ovarian cancer patients
Scientific Reports (2021)