Abstract
The intestinal parasite, Cryptosporidium, is a major contributor to global child mortality and causes opportunistic infection in immune deficient individuals. Innate resistance to Cryptosporidium, which specifically invades enterocytes, is dependent on the production of IFN-γ, yet whether enterocytes contribute to parasite control is poorly understood. In this study, utilizing a mouse-adapted strain of C. parvum, we show that epithelial-derived IL-18 synergized with IL-12 to stimulate innate lymphoid cell (ILC) production of IFN-γ required for early parasite control. The loss of IFN-γ-mediated STAT1 signaling in enterocytes, but not dendritic cells or macrophages, antagonized early parasite control. Transcriptional profiling of enterocytes from infected mice identified an IFN-γ signature and enrichment of the anti-microbial effectors IDO, GBP, and IRG. Deletion experiments identified a role for Irgm1/m3 in parasite control. Thus, enterocytes promote ILC production of IFN-γ that acts on enterocytes to restrict the growth of Cryptosporidium.
Similar content being viewed by others
Introduction
The intestinal epithelium is an important site for nutrient uptake and a barrier to micro-organisms. However, this barrier can be disrupted by a diverse group of viruses, bacteria and parasites that infect the gastrointestinal tract. The enteric diseases caused by these pathogens are of public health importance, accounting for 8–10% of deaths in children worldwide.1 While many of these pathogens will disseminate from the gut, a subset is restricted to the epithelial layer, where their interactions with enterocytes are likely key determinants of disease outcome. The epithelium is composed of enterocytes, a large population of columnar epithelial cells necessary for nutrient uptake, as well as subsets of specialized epithelial cells including Paneth, goblet, and tuft cells that have roles in mucosal homeostasis and immune defense.2,3,4,5,6,7,8,9,10
How enterocytes participate in resistance to different types of infection is a fundamental question that is particularly relevant to epithelial-restricted pathogens such as rotavirus, norovirus, astrovirus, Shigella, Cyclospora, and Cryptosporidium. The apicomplexan parasite, Cryptosporidium, is a leading cause of severe diarrhea and death in infants11,12,13,14 and a common opportunistic infection in individuals with primary and acquired immune deficiencies.15,16 Currently, there are no fully effective drugs or vaccines to treat or prevent cryptosporidiosis. Based on clinical experience and animal models, effective control and clearance of Cryptosporidium is dependent on cell-mediated immunity and the production of IFN-γ.17,18,19,20 Cryptosporidium invasive stages infect the apical surface of enterocytes, where they transform into the replicative trophozoite and occupy a unique intracellular but extracytoplasmic niche.21 The restriction of this parasite to the intestinal epithelium provides a model to understand how the immune system senses enteric pathogens and an opportunity to identify enterocyte-driven pathways that promote the control of intracellular pathogens.
Most human infections are caused by the anthroponotic Cryptosporidium hominis and the zoonotically transmitted C. parvum. C. parvum is the species most widely used as an experimental model, however it does not robustly infect adult immune competent mice. For this reason, most studies with this organism have used neonates or immune deficient mice (such as those that lack IFN-γ or components of adaptive immunity) to help define important pathways for resistance to Cryptosporidium. This approach has provided evidence that dendritic cell (DC) derived IL-12 promotes NK cell production of IFN-γ required for innate restriction of this infection.22,23,24,25,26 IL-18 is another cytokine that promotes resistance to C. parvum, but it is unclear if this protection is due to its ability to enhance ILC production of IFN-γ or whether IL-18 directly activates epithelial cells to limit parasite growth.27,28,29,30 Likewise, while IFN-γ is important for parasite control, multiple cell types in the gut can respond to this cytokine and, thus, it is unclear whether the protective effects of IFN-γ are mediated through enterocytes, DCs, or macrophages.
Recent studies have developed the means to genetically modify C. parvum that is dependent on the ability to select for transgenic parasites while being passaged in IFN-γ-deficient mice.31 Over time, this has resulted in parasite adaptation whereby these transgenic C. parvum readily infect wild-type (WT) mice and provide an experimental system to dissect the events required for protective immunity in an immunocompetent setting. In this study, infection with mouse-adapted C. parvum (maCp) revealed that intestinal type-1 innate lymphoid cells (ILC1s) are a rapid source of IFN-γ that limits parasite growth. This protective response was dependent on the production of IL-12 and epithelial-derived IL-18. Lineage-specific deletion of STAT1, a critical transcription factor downstream of IFN-γ signaling, demonstrated that STAT1 was uniquely required in enterocytes—but not DCs or macrophages—to restrict parasite growth. Transcriptional profiling of enterocytes from infected mice highlighted an IFN-γ signature, and subsequent deletion experiments identified immune-related GTPase-m1 and -m3 (Irgm1/3) as downstream effectors of the IFN-γ-mediated response. Together, these studies establish that enterocytes have a central role as a source of IL-18 required to stimulate local ILC responses and are key mediators of IFN-γ-mediated protection to an important enteric pathogen.
Results
Localized innate IFN-γ provides early constraint of Cryptosporidium infection
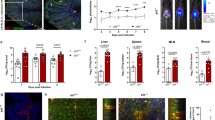
To assess the relationship between parasite burden and IFN-γ production, WT mice treated with an isotype control antibody (IgG) or anti-IFN-γ (α-IFN-γ) were infected with a mouse-adapted C. parvum strain that expressed nanoluciferase (maCp-Nluc).31,32 At 4 days post-infection (dpi), paired biopsies were taken along the entirety of the intestine and used to quantify nanoluciferase activity or placed in culture to assess levels of secreted IFN-γ. In control mice, parasite replication was readily detected and restricted to the distal small intestine but levels of IFN-γ in ileal supernatants were not elevated above uninfected controls (Fig. 1a). However, in mice treated with anti-IFN-γ, there was an 80-fold increase in parasite burden and ex vivo IFN-γ was readily detected and correlated with areas of the gut with the highest parasite burdens (Fig. 1a, filled circles). Thus, the early production of IFN-γ provides a mechanism of resistance to C. parvum, but the ability to detect the infection-induced production of IFN-γ is dependent on the presence of a high tissue parasite burden.
WT mice were infected with 105 maCp-Nluc oocysts and treated with either α-IFN-γ or isotype control. a At 4 dpi, two adjacent 5 mm biopsies were taken along the length of the small and large intestines (D duodenum, J jejunum, I ileum, Ce cecum, and C colon). One biopsy was evaluated for nanoluciferase (top), the other was incubated at 37 °C for 24 h and IFN-γ was measured from the supernatant. Graph depicts 1 representative mouse from n = 3 for each group. b WT C57BL/6 and Rag2−/− mice were treated with isotype or α-IFN-γ and nanoluciferase was used to measure fecal oocyst shedding, n = 4 per group. Similar results were observed in two additional experiments. c Representative mucosa and (d–g) cumulative histology scoring from Rag2−/− mice 5 dpi that shows a marked increase in cryptosporidia organisms (black arrows) infecting the villus enterocytes in treated mice versus controls with a progressive increase in epithelial dysplasia (arrowheads), reduced villus:crypt ratios with increased crypt depth (double-headed arrows) and crypt mitoses (white arrows); scale bars = 20 μm. h Rag2−/− mice were infected with 105 maCp-Nluc oocysts and treated with α-IFN-γ at 50, 53, and 56 dpi. Oocysts shedding was monitored throughout by nanoluciferase. n = 4, representative from three experimental replicates. Bars denote mean ± SD. ANOVA followed by multiple comparisons were performed on cumulative pathology scores, **p ≤ 0.01; ***p ≤ 0.001.
To assess the contribution of innate and adaptive sources of IFN-γ on early resistance to maCp-Nluc, WT, and Rag2−/− mice (lacking T and B cells) were treated with α-IFN-γ or an isotype control prior to infection. No gross histological differences were detected between mouse strains at steady state (Supplementary Fig. 1A). In both WT and Rag2−/− mice, IFN-γ neutralization resulted in increased oocyst shedding by 4 dpi, which was exacerbated at 6 dpi (43-fold in WT and 263-fold in Rag2−/−, Fig. 1b), and correlated with histological analysis that showed enhanced parasite burden in Rag2−/− mice treated with α-IFN-γ (Fig. 1c, d). In addition, there were significant infection-induced local changes in mucosal architecture and enterocyte morphology by 5 dpi, including reduced average villus:crypt ratio, increased crypt mitoses and a trend toward increased epithelial dysplasia (Fig. 1e–g). However, quantification of these changes across multiple sections did not yield statistically significant changes between infected mice and those treated with α-IFN-γ. However, when observing ileal pathology at a later timepoint, α-IFN-γ treatment resulted in more severe pathological changes (12 dpi Supplementary Fig. 1B). This pathology corresponds with the sustained oocyst shedding observed after the peak parasite burden at 6 dpi. Interestingly, treatment of chronically infected Rag2−/− mice with α-IFN-γ at 50 dpi led to a marked recrudescence in parasite burden (Fig. 1h). These data identify an innate mechanism of IFN-γ-mediated resistance to C. parvum that operates during the acute and chronic phases of infection, but which is insufficient for parasite clearance.
Innate lymphoid cells are required for control of Cryptosporidium
There are several ILC populations capable of IFN-γ production that include: NK cells, ILC1s and ILC3s. Early studies with C. parvum concluded that NK cells were a major source of IFN-γ, but the use of α-asialo-GM1 (which depletes NK cells but not other ILC populations) indicated that there may be other innate sources of IFN-γ.26,33 Indeed, when Rag2−/− mice infected with maCp-Nluc were treated with α-asialo-GM1, there was efficient depletion of splenic NK cells but the intestinal ILCs (CD45.2+ NK1.1+NKp46+) and levels of secreted IFN-γ were only reduced by approximately half (Supplementary Fig. 1C–E). Therefore, to assess the role of ILCs in resistance to C. parvum, Rag2−/−Il2rg−/− mice, which lack all ILCs in addition to T and B cells, were compared to WT and Rag2−/− mice. After infection with maCp-Nluc, oocyst shedding declined in WT mice by 9 dpi, and resolved by 18 dpi (Fig. 2a). Parasite levels in Rag2−/− mice were not elevated compared to WT mice at early time points (3–9 dpi), but the infection failed to resolve in the absence of adaptive immune cells. In contrast, Rag2−/−/Il2rg−/− mice demonstrated little evidence of parasite control at any time examined, with ~250-fold higher nanoluciferase readings by 6 dpi, compared to Rag2−/− mice, which were sustained for the duration of the experiment (Fig. 2a). In uninfected mice, there were no gross histological differences in the ileal tissues among the three strains (Supplementary Fig. 1A). At 5 dpi, consistent with the data in Fig. 1b, WT and Rag2−/− mice had similar numbers of parasites and levels of infection-induced changes to the epithelium (Fig. 2d–g, white and gray bars). In contrast, Rag2−/−/Il2rg−/− mice exhibited dramatically increased parasite burden (Fig. 2b, c). The absence of ILCs also resulted in increased mitoses and crypt branching, as well as more severe villus pathology, indicated by increased epithelial dysplasia and attenuation (Fig. 2d–g, black bars). Thus, in the absence of adaptive immunity, ILCs are required for both early and long-term control of Cryptosporidium and the lack of these cells results in unrestricted parasite replication and severe intestinal pathology.
C57BL/6 WT, Rag2−/−, and Rag2−/−Il2rg−/− mice were infected and used for histological analysis or oocyst shedding. a Kinetics of fecal oocyst shedding; n = 3–5, representative of three replicates comparing Rag2−/− and Rag2−/−Il2rg−/−. b–g H&E staining of representative villi (upper panels) and crypts (lower panels), and cumulative histology scoring from WT, Rag2−/− and Rag2−/−Il2rg−/− mice at 5 dpi that show a marked increase in Cryptosporidium organisms (black arrows) infecting the villus enterocytes in Rag2−/−Il2rg−/− mice versus WT or Rag2−/− mice as well as progressive villus epithelial dysplasia with attenuation (arrowheads), increases in crypt branching (double arrows) and crypt mitoses (blue arrows); scale bars = 20 μm; n = 4–8 where each symbol denotes one mouse cumulative of three experiments. Bars indicate mean ± SD. n = 4–8 where each symbol denotes one mouse, cumulative of three experiments. Bars indicate mean ± SD. For histology scoring, t tests were used to compare uninfected and infected within each mouse strain, *p ≤ 0.05; **p ≤ 0.01; ****p ≤ 0.0001.
ILC1s are a major source of innate IFN-γ during C. parvum infection
To determine the innate cellular source(s) of IFN-γ in the intestinal epithelium, mice in which the gene for the surface-expressed protein, Thy1.1, is under the transcriptional control of the Ifng promotor34 were infected with mCherry-expressing maCp-Nluc. This approach allowed the simultaneous quantification of parasite-infected (mCherry+) enterocytes (EpCAM+CD45−) and ILCs (CD45.2+CD3−CD19−NK1.1+NKp46+) producing IFN-γ (Thy1.1+). Based on the data in Fig. 1a, groups of mice were also treated with α-IFN-γ to increase the ability to detect cells that produce IFN-γ. In uninfected mice, enterocytes lack mCherry, and a low basal percentage of ILCs express surface Thy1.1 regardless of α-IFN-γ treatment (Fig. 3a, c). At 4 dpi, a detectable portion of IECs were infected (mCherry+), and there was a small increase in the frequency of Thy1.1+ ILCs. Treatment with α-IFN-γ resulted in a significant increase in parasite burden (Fig. 3a, b) and a 3–5-fold increase in the proportion of Thy1.1+ ILCs (Fig. 3c, d). This increased production of IFN-γ was specific to the intestinal epithelium, as few cells expressed Thy1.1 in Peyer’s patches or mesenteric lymph nodes, even with IFN-γ neutralization (Fig. 3e, f). Of note, in these infected immune competent reporter mice, myeloid cells (EpCAM−CD3−CD19−NK1.1−) did not express Thy1.1 (data not shown), but a proportion of intestinal CD4+ T cells and, to a lesser extent, CD4+CD8α+ T cells did express increased levels of Thy1.1 by 5 dpi (Supplementary Fig. 2A).
Ifng−Thy1.1 KI mice were infected with mCherry-expressing maCp-Nluc and cells from the ileal epithelium were assessed by flow cytometry at 4 dpi. a Representative flow plots and (b) summary bar graph of maCp-infected (mCherry+) intestinal epithelial cells, gated on Live CD45.2−EpCAM+ cells. c Representative flow plots and (d) summary bar graph of Ifng+ (Thy1.1+) innate lymphoid cells (NK1.1+NKp46+), n = 7–24 from four experiments. e Representative flow plots and (f) summary bar graph of frequency of Ifng+ (Thy1.1+) cells from ILCs (seeing gating strategy from (c)) taken from intestinal epithelium (IE), Peyer’s patches (PP), or mesenteric lymph nodes (mesLN). mesLN and PP were pooled from 2 to 4 mice, representative of three experiments. g Representative flow plots and (h) summary bar graph of T-bet and Eomes expression on Ifng+ (Thy1.1+) or Ifng− (Thy1.1−) ILCs. i Representative flow plots and (j) summary bar graph of T-bet and RORγt expression on Ifng+ (Thy1.1+) or Ifng− (Thy1.1−) ILCs. All ILCs were gated on Live CD45.2+CD3−CD19−NKp46+NK1.1+ cells. Statistical significance between α-IFN-γ treated or untreated in (b, d) was determined by Student’s t test. Statistical significance in (f) was determined by one-way ANOVA and multiple comparisions. Student’s t tests were used to compare Thy1.1+ and Thy1.1− in (h, j). ns not significant (p > 0.05), *p ≤ 0.05; **p ≤ 0.01; ****p ≤ 0.0001.
To characterize the ILC subsets that produce IFN-γ, expression of the transcription factors Eomes, T-bet and RORγt was used to distinguish NK cells, ILC1s, and ILC3s, respectively. In uninfected reporter mice, the ILC populations in the gut were composed of 50–60% T-bet+ ILC1s, 30–40% Eomes+ NK cells and 5–10% were RORγt+ ILC3s (Supplementary Fig. 2B). All three of these ILC subsets were also present at 4 dpi (Fig. 3g, i). Among ILCs not expressing IFN-γ (Thy1.1−), the proportions of NK cells, ILC1s and ILC3s were similar to those of uninfected mice (Fig. 3g, i, bottom plots and Fig. 3h, j open circles). However, in infected mice, the proportion of T-bet+ ILCs was significantly increased in cells expressing Ifng (Thy1.1+), where ~80% of the Thy1.1+ cells expressed T-bet and not Eomes or RORγt (Fig. 3g–j, filled circles). In addition, while NK cells in the mesenteric lymph nodes expressed CD27 and CD49b, the Thy1.1+ cells in the intestinal epithelium showed low expression of these NK cell markers (Supplementary Fig. 2C, D). These data indicate that within days of Cryptosporidium infection, ILC1s present in the intestine become activated and are a major source of IFN-γ.
Epithelial-derived IL-18 synergizes with IL-12 to promote ILC production of IFN-γ
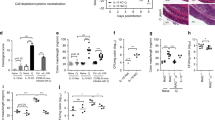
IL-12 and IL-18 have roles in innate resistance to Cryptosporidium sp.22,27,32 and can, in other experimental systems, synergize to stimulate ILC production of IFN-γ.35,36,37 To evaluate their contributions to early resistance to C. parvum, the level of oocyst shedding in WT, Ifng−/−, Il12b−/− (IL-12p40), and Il18−/− mice were compared. As expected, infection was established in WT mice, and Ifng−/− mice showed a rapid and marked increase in parasite burden. While the parasite levels in Il12b−/− mice were comparable to Ifng−/− mice, Il18−/− mice showed a phenotype that was intermediate between WT and Ifng–/– mice (Fig. 4a). A recent study highlighted BATF3-dependent CD103+ DCs as a source of IL-12 during neonatal infection with C. parvum,24 but the cellular source of IL-18 required to control Cryptosporidium was unclear. To determine the importance of the intestinal epithelium as a cellular source of IL-18 during cryptosporidiosis, oocyst shedding from mice bearing an epithelial lineage-specific deletion of IL-18 (Vil1-Cre × Il18fl/fl, here referred to as Il18ΔIEC) was compared to WT and Il18−/− mice. In these studies, the levels of infection in Il18ΔIEC mice were comparable to whole-body Il18−/− mice (Fig. 4b), suggesting that the intestinal epithelium is a key source of the IL-18 that mediates early resistance to C. parvum.
a WT C57BL/6 mice or mice lacking IL-18 (Il18−/−), IL-12p40 (Il12b−/−), or IFN-γ (Ifng−/−) were infected with maCp-Nluc and oocyst shedding in feces was monitored. Representative of three experimental replicates. b Control (Cre−Il18fl/fl), epithelial-specific IL-18-deficient (Vil1-Cre × Il18fl/fl Il18ΔIEC) and Il18−/− mice were infected with maCp-Nluc and oocyst shedding in feces was monitored. c–g Rag2−/− mice (c) or Ifng-Thy1.1 KI mice (d–g) were treated with α-IL-18, α-IL-12p40, or both and infected with maCp-Nluc. c Oocyst shedding in feces was monitored. d Representative flow plots and (e) summary bar graph of maCp-infected (mCherry+) IECs 4 dpi. f Representative flow plots and (g) summary bar graph of Ifng+ (Thy1.1+) ILCs 4 dpi (gating same as Fig. 3). n = 8–11 from three experiments. Statistical significance was determined by one-way ANOVA and multiple comparisions; ns not significant (p > 0.05) ****p ≤ 0.0001.
The experiments that utilized mice with germline deletion of IL-18 or IL-12 identified an important role for these cytokines in early resistance to C. parvum. However, analyses of naive Il12b−/− mice revealed a marked absence of ILC populations in the intestinal epithelium (Supplementary Fig. 3A–C) and previous studies have reported that germline deletion of IL-18 impacts immune homeostasis in the gut.38 In order to control for these confounding effects, C57BL/6 and Rag2−/− mice were treated with antibodies against IL-12p40, IL-18, or both beginning 4 days prior to infection with maCp-Nluc. In WT and Rag2−/− mice, neither treatment alone dramatically impacted susceptibility to maCp-Nluc but the simultaneous blockade of IL-12 and IL-18 led to a 14.5-fold increase in parasite burden (Rag2:−/− Fig. 4c, BL/6: Supplementary Fig. 3D). Moreover, despite having similar infection burdens to α-IFN-γ treated mice (Fig. 4d, e), Ifng-Thy1.1 reporter mice treated with α-IL-12p40 plus α-IL-18 showed nearly complete loss of Thy1.1+ ILC (Fig. 4f, g). Collectively, these experiments demonstrate that epithelial-derived IL-18 synergizes with IL-12 to stimulate ILC production of IFN-γ required for early restriction of Cryptosporidium infection.
Enterocytes are critical contributors to early IFN-γ-mediated resistance to C. parvum
While IFN-γ is important in acute resistance to Cryptosporidium, it is unclear whether it acts directly on infected cells to restrict parasite growth, or indirectly via activation and maturation of macrophages and DCs.39 The transcription factor STAT1 is the major mediator of IFN signaling. Ifng−/− and Stat1−/− mice had similarly enhanced susceptibility to maCp, suggesting that IFN-γ is the main driver of STAT1 signaling required for resistance to this parasite (Supplementary Fig. 4A). Therefore, lineage-specific Cre-lox mediated deletion was utilized to identify cell subsets in which STAT1 signaling was critical for early control of maCp. Loss of STAT1 only in DCs (Cd11c-Cre × Stat1;fl/fl Stat1ΔDC) or macrophages (Lyz2-Cre × Stat1;fl/fl Stat1ΔMΦ) did not enhance parasite burden over Cre− controls. In contrast, when STAT1 was deleted from enterocytes using a tamoxifen-inducible deletion (Vil1-CreERT2 × Stat1;fl/fl Stat1ΔIEC), oocyst shedding was greatly increased (Fig. 5a) and at a level that paralleled complete Stat1−/− mice (Fig. 5b). This was not due to reduced levels of IFN-γ in the intestine (Supplementary Fig. 4B). These data indicate that IFN-γ induced STAT1-mediated activity in intestinal epithelial cells provides a cell-intrinsic mechanism for the control of Cryptosporidium.
a, b Cre−Stat1fl/fl (Control), Cd11c-Cre × Stat1fl/fl (Stat1ΔDC), and Lyz2-Cre × Stat1fl/fl (Stat1ΔMΦ), Vil1-CreERT2-CreERT2 × Stat1fl/fl (Stat1ΔIEC), and Stat1−/− mice were infected with maCp-Nluc and oocyst shedding was monitored in feces. c–f IEC from naïve mice or mice infected with maCp-mCherry for 3 or 5 days were FACS sorted and used for RNA-sequencing analysis. c Volcano plot with IFN signature genes marked in red. DEGs that were significantly upregulated (p ≤ 0.05) in IECs from maCp-mCherry-infected mice are labeled. d, e Gene set enrichment analysis highlighting an IFN-γ signature (d), a cluster of IFN pathways (e). f Heatmap of pertinent IFN-stimulated genes. g WT C57BL/6, Ido1−/−, GBPchr3, and Irgm1/m3−/− mice were infected with 5 × 104 maCp-Nluc oocysts and fecal shedding of oocysts was monitored. Representative of three experimental replicates.
Because the mechanism that leads to the IFN-γ-mediated restriction of Cryptosporidium in enterocytes is unknown, transcriptional profiling of sort-purified enterocytes from uninfected or infected mice was performed. RNA-seq analysis of enterocytes identified a limited set of differentially expressed genes; that included several IFN-inducible GTPases: Igtp (encoding Irgm3), Iigp1 (encoding Irga6), Gm12250 (encoding Irgb10), Gbp7, Tgtp1, Ifi47 (encoding Irg-47), and Irgm1 (Fig. 5c). Despite the limited number of differentially expressed genes, GSEA showed a strong enrichment of IFN-γ signaling in enterocytes from infected mice (Fig. 5d), consistent with expression of transcripts for Ifngr1 and Ifngr2 in IECs (data not shown). Enrichment mapping software was used to define related gene functions and examine overlap between enriched gene sets. This analysis identified three main clusters: IFN signatures, mitochondrial signatures, and protein translation/ribosome signatures (Fig. 5e and Supplementary Fig. 4C, D). The mitochondrial and ribosome signatures have been associated with enterocyte stress40,41 and likely reflect the increased mitotic index and epithelial dysplasia associated with this infection (see Figs. 1, 2). Furthermore, while gene sets encompassing both IFN-γ and type I IFNs were enriched, the degree of overlap was greatest among the all-encompassing “interferon signaling” and the two interferon gamma gene sets, further indicating that IFN-γ signaling was the dominant response in infected IECs.
Given the dominant IFN-γ signature, the expression data sets were curated for changes in IFN-γ-induced genes associated with control of intracellular pathogens (Fig. 5f). Genes strongly upregulated by maCp infection included those encoding for β2m and CIITA (Class II Major Histocompatibility Complex Transactivator) which affect MHC I and MHC II expression, respectively (Fig. 5f). Also markedly upregulated were transcripts encoding for indolamine dioxygenase (IDO), several Guanylate Binding Proteins (GBPs), and Immunity Related GTPases (IRGs), all of which are known to be important in non-hematopoietic cells for IFN-γ to restrict the growth of T. gondii.42,43 However, their roles in resistance to Cryptosporidium have not been established. To test the impact of these pathways on maCp, the course of infection in Ido1−/−, GBPchr3 (lacking Gbp2, Gbp3, Gbp5, Gbp7, and Gbp2ps) and Irgm1/m3−/− mice was compared with that in WT mice by fecal oocyst shedding. Because Irgm1-deficient mice have multiple defects in immune function that are mitigated by the loss of Irgm3, Irgm1/m3−/− mice are used to study the role of Irgm1 in resistance to infections.44,45 While Ido1−/− and GBPchr3 mice showed no enhanced susceptibility to maCp-Nluc, Irgm1/m3−/− mice demonstrated a greater than fivefold increase in fecal oocyst shedding at 5 dpi (Fig. 5g). We note that parasite burden in Irgm1/m3–/– mice was intermediate between WT and Ifng−/− or Stat1ΔIEC mice, which argues for additional IFN-γ/STAT1-mediated mechanisms for parasite control. These findings provide the first evidence for a role of IRGs in the IFN-γ-mediated mechanism of resistance to Cryptosporidium.
Discussion
The importance of IFN-γ in resistance to Cryptosporidium is well appreciated,17,19,46 but significant gaps exist in our understanding of the specific cells that produce and respond to this central cytokine in order to mediate parasite control. Previous studies have focused on NK cells as a potential innate source of IFN-γ in resistance to C. parvum;25,26 however, they predate the identification of other ILC populations. As confirmed in our studies, NK cells are largely present in secondary lymphoid tissues and the circulation, whereas the other ILC subsets are predominately tissue resident.47 Although there is marked phenotypic and functional plasticity for ILCs in the intestine,48 our studies with mouse-adapted C. parvum suggested that ILC1s were the major early source of IFN-γ in the small intestine, although NK cells likely also contributed. In other models of intracellular infection, NK and ILC1-mediated resistance is transient, associated with the acute phase of infection but not sufficient for long-term control.49,50 Thus, it was unexpected that although innate production of IFN-γ was not sufficient for parasite clearance in Rag−/− mice, it did provide a significant level of long-term control of maCp. Because of the lack of effective therapies to treat chronic cryptosporidiosis in patients with primary or acquired defects in T-cell function,51,52,53 this model provides an opportunity to understand how ILC responses are maintained and whether they can be enhanced to mediate parasite clearance.
Previous studies have demonstrated that IL-12 and IL-18 are important in resistance to C. parvum.20,22,27,29,54 While DCs are a major source of IL-12,24,55,56 a recent study reported that, in vitro, DCs incubated with C. parvum activate the inflammasome and downstream caspase-1, resulting in IL-18 secretion.30 In addition, IL-18 has been proposed to promote parasite control independent of IFN-γ in enterocyte cell lines.29 In contrast, we found that enterocyte-intrinsic NLRP6 inflammasome activation was required for IL-18 mediated resistance to Cryptosporidium in vivo.32 Consistent with that observation, the studies presented here highlight enterocytes as a critical source of IL-18 which synergized with IL-12 to stimulate ILC1s to produce early IFN-γ. During infection with Citrobacter rodentium, an extracellular bacteria that attaches to the luminal surface of enterocytes, NLRP3 inflammasome activation leads to enterocyte-derived IL-18, which is required for host resistance.57,58,59 Thus, different sensors in enterocytes allow these cells to respond to diverse pathogens but converge on the processing of IL-18. In contrast, although Salmonella infects enterocytes, enteric neurons (not enterocytes) are the relevant source of IL-18 required for protection.60 Intriguingly, patients with Hirschsprung disease have regions that lack distal bowel ganglia and are susceptible to Cryptosporidium,61,62 but additional experiments will be required to determine whether the enteric nervous system is also a relevant source of IL-18 required for resistance to Cryptosporidium.
IFN-γ is a cytokine with wide ranging effects relevant to Cryptosporidium that include its ability to enhance antigen presentation and activation of anti-microbial activities within non-hematopoietic cells. Here, deletion of STAT1 was utilized to target the downstream effects of IFN-γ during maCp infection but it is important to note that STAT1 is utilized by other IFNs and cytokines such as IL-27. Indeed, there are reports that endogenous type I63 and type III IFNs64 contribute to early resistance to C. parvum. However, we have not observed a role for IL-27 or type I IFNs in parasite control (unpublished observations). Nevertheless, in the current study, mice deficient in STAT1 or IFN-γ showed similar susceptibility and the genes most strongly enriched in enterocytes from infected mice are more closely associated with IFN-γ, compared to IFN-α/β or IFN-λ.65 The finding that the loss of STAT1 in macrophage and DC populations did not impact susceptibility indicated that these populations were not important effectors of IFN-γ-mediated parasite control. In contrast, the inducible deletion of STAT1 in enterocytes established a cell-intrinsic role for STAT1 in resistance to C. parvum. This pathway contrasts with Salmonella, where IFN-γ is required to restrict bacterial growth in macrophages but not enterocytes,66,67 as well as Toxoplasma gondii, where innate sources of IFN-γ act specifically on cDC1s.68
Studies using human intestinal cell lines concluded that IFN-γ can inhibit growth of Cryptosporidium, but inhibitory effects in vitro are modest and the mechanisms that underlie parasite restriction are not understood.14,46 The identification of a role for Irgm1 suggests a potential overlap with mechanisms used to control other intracellular pathogens. The upregulation of Irgm1, Igtp, and Iigp1 in enterocytes from infected mice highlights anti-microbial effectors that, together with the GBPs, can intersect with autophagy pathways involved in pathogen restriction.69 It is important to note that Irgm1 and Irgm3 are also expressed in other cell types, thus the use of germline deletion does not rule out additional roles for these proteins in resistance to C. parvum. Furthermore, while the loss of the GBPs on chromosome 3 (GBP 1, GBP2, GBP3, GBP5, and GBP7) had no detectable impact on host susceptibility, it does not rule out possible roles for the other GBPs located on chromosome 5. Of these, the expression of Gbp6 was significantly enriched in epithelial cells from infected mice. Another possibility is that, although GBPs interact with vacuoles that contain other intracellular pathogens, perhaps the extracytoplasmic location of Cryptosporidium and the partitioning by parasite induced cellular barriers precludes this interaction,21 which would render the canonical GBP-dependent mechanisms of protection ineffective. The recent advances using enteroid-derived systems to culture Cryptosporidium70,71 should be useful to dissect how enterocytes utilize interferon-stimulated genes to directly clear or restrict growth of Cryptosporidium species.
The intestinal epithelium is a critical barrier where the immune system interacts with diverse microbial communities in the gut. Enterocyte interactions with the microbiome are central to homeostasis and the etiology of a variety of allergies and inflammatory conditions.5,72,73 There is an increased appreciation that different types of epithelial cells provide signals that help to coordinate the enteric nervous system, gut physiology and mucosal immunity in order to maintain tolerance. For extracellular pathogens, enterocytes promote clearance of the helminth Trichuris muris74 and the bacteria Clostridium difficile75 and Citrobacter rodentium.57 Less is known about how enterocytes respond to intracellular infections, although inflammasome-mediated extrusion of Salmonella-infected enterocytes limits bacterial spread.76 Indeed, expulsion of infected enterocytes represents a conserved mechanism of pathogen resistance that is present in insects and higher vertebrates.77,78,79 The studies presented here identified enterocytes as a critical source of IL-18 required for ILC activation and as key drivers of IFN-γ mediated resistance to an important enteric pathogen. Thus, mouse-adapted C. parvum is a valuable model of an enteric infection that affects intestinal physiology and nutritional status. Furthermore, the ability to genetically modify this organism provides the opportunity to understand the role of enterocytes in pathogen recognition and resistance to infection.
Materials and methods
Mice
C57BL/6 (stock no: B6NTac), Rag2−/− (stock no: RAGN12), and Rag2−/−Il2rc−/− (stock no: 4111) were purchased from Taconic. C57BL/6 (stock no: 000664), Rag2−/− (stock no: 008449), Ifng−/− (stock no: 002287), Il12rb−/− (stock no: 002693), Stat1−/− (stock no: 012606), Il18−/− (stock no: 004130), Ido1−/− (stock no:005867), Vil1-Cre (stock no:021504), Lyz2-Cre (stock no: 004781), and Cd11c-Cre (stock no. 008068) were purchased from Jackson Laboratory and maintained in-house. Stat1flox mice were generated as previously described80 and maintained in house. Ifng/Thy1.1 BAC-In mice were provided by Dr. Phillip Scott but originated in the laboratory of Dr. Casey Weaver.34,81 Il18flox mice were provided by Dr. Jorge Henoa-Mejia. Vil1-CreERT2 were provided by Dr. Lou Ghanem at Children’s Hospital of Philadelphia and Dr. David Artis at Cornell University. In-house breeding was performed to obtain all Cre-lox combinations. Unless otherwise noted, mice used in this study were males or females ranging from 7 to 11 weeks. We did not observe a difference in infection burden between male and female mice. All mice were age matched within individual experiments. All protocols for animal care were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania (protocol #805405 and #806292).
Parasites and infection
Transgenic C. parvum expressing nanoluciferase and mCherry32,82 are propagated by orally infecting Ifng−/− mice. Oocysts are purified from fecal collections of infected mice using sucrose flotation followed by a cesium chloride gradient, as previously described.82 Mice were infected with 5 × 104–1 × 105 oocysts by oral gavage. To measure parasite burden in intestinal tissue, 5 mm biopsy punches were taken along murine small intestines and suspended in 0.5 mL lysis buffer (50 mM tris HCl (pH 7.6), 2 mM DTT, 2 mM EDTA, 10% glycerol, and 1% TritonX in ddH2O). To quantify fecal oocyst shedding, 20 mg fecal material was suspended in 1 mL lysis buffer. Samples were shaken with glass beads for 5 min, then combined in a 1:1 ratio with Nano-Glo® Luciferase solution (Promega, Ref N1150). A Promega GloMax plate reader was used to measure luminescence. For each experiment, comparators are described in the text and involve cage-to-cage comparisons. The numbers of mice per experimental group and experimental replicates are provided in the figure legends and no data sets were excluded. No randomization or blinding strategies were employed.
Histology
For histological analysis of the small intestine, tissue from the distal third of the small intestine was flushed with 10% neutral buffered formalin (Sigma, St Louis, MO, USA), then “swiss-rolled” and fixed overnight. Fixed samples were paraffin-embedded, sectioned, and stained with hematoxylin and eosin for detailed histologic evaluation. Slides were evaluated by a board-certified veterinary pathologist in a blinded fashion for quantitative measurements of number of parasites, villus/crypt architectural features and inflammatory infiltrates, and semiquantitative scores for villus epithelium lesions as previously described.82
Cytokine neutralization and measurement
To neutralize IFN-γ, 1 mg anti-IFN-γ (XMG1.2, BioXcell Cat #: BE0055) was given intraperitoneally (i.p.) 1 day prior and 2 days post-infection with maCp. To also neutralize IL−12 or IL-18, 2 mg anti-IL-12p40 (C17.8 BioXcell Cat #: BE0051) or 1 mg anti-IL-18 (YIGIF74-1G7, BioXcell Cat #: BE0237) were given i.p. on days −4, −1, and 1, while anti-IFN-γ was given on days −2 and day 2. Intestinal IFN-γ levels were assessed from 5 mm biopsy punches that were incubated in complete RPMI at 37 °C for 24 h. Clear, flat-bottom 96-well plates (Immunulon 4 HBX) were coated with 0.25 μg/mL anti-IFN-γ (AN-18, Invitrogen) at 4 °C overnight. Samples were added and IFN-γ left to bind at 37 °C for 2 h. 0.25 μg/mL biotinylated anti-IFN-γ (R4–6A2, eBioscience) in PBS with 2.5% FBS and 0.05% Tween was added for 1 h at room temperature, followed by peroxidase-labeled streptavidin for 30 min. Finally, KPL ABTS® peroxidase substrate (SeraCare Cat #: 5120-0041) was applied for detection.
Flow cytometry and cell sorting
Single-cell suspensions were prepared from intestinal sections by shaking diced tissue at 37 °C for 25 min in Hank’s Balanced Salt Solution with 5 mM EDTA and 1 mM DTT. Cell pellets were then passed through 70 mm and 40 mm filters. Cells were surface stained using the following fluorochrome-conjugated Abs: anti-EpCAM (G8.8), anti-CD45.2 (104), anti-CD19 (MB19-1), anti-Thy1.1 (HIS51), anti-CD27 (LG.7F9), anti-RORγt (B2D) and anti-γδ TCR (eBioGL3) from eBioscience; anti-NK1.1 (PK136), anti-NKp46 (29A1.4), anti-CD3 (17A2), anti-CD127 (SB/199), anti-CD49a (HMα1), anti-T-bet (4B10), and anti-CD8β (YTS156.7.7) from BioLegend; anti-Eomes (Dan11mag) and Live/Dead Aqua from Invitrogen; and anti-CD49b (DX5) from BD Biosciences. Data were collected on a LSRFortessa (BD Biosciences) and analyzed with FlowJo v10 software (TreeStar). Cell sorting was performed on a BD FACS Jazz (BD Biosciences).
RNA-seq and gene enrichment analysis
RNA-seq reads were pseudo-aligned to the Ensembl Mus musculus reference transcriptome v79 using Kallisto v0.44.0.83 In R, transcripts were collapsed to genes using Bioconductor tximport,84 and differentially expressed genes were identified using Limma-Voom.85,86 Gene set enrichment analysis was performed using the GSEA software and the annotated gene sets of the Molecular Signatures Database (MSigDB).87,88 From the GSEA output, enrichment maps were generated to provide a visual representation of gene set overlap using Cytoscape v3.8.2.89 Data and analyses have been deposited to the GEO repository (GSE168680). One sample was excluded for failing quality control.
Statistics
Statistical significance was calculated using the unpaired Student’s t test for comparing two groups, or ANOVA followed by multiple comparisons for comparing groups of three or more. Analyses were performed using GraphPad Prism v.9.
References
Collaborators GBDCM. Global, regional, national, and selected subnational levels of stillbirths, neonatal, infant, and under-5 mortality, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1725–1774 (2016).
Birchenough, G. M., Johansson, M. E., Gustafsson, J. K., Bergstrom, J. H. & Hansson, G. C. New developments in goblet cell mucus secretion and function. Mucosal Immunol. 8, 712–719 (2015).
von Moltke, J., Ji, M., Liang, H. E. & Locksley, R. M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225 (2016).
Adolph, T. E. et al. Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272–276 (2013).
Peterson, L. W. & Artis, D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14, 141–153 (2014).
Cliffe, L. J., Potten, C. S., Booth, C. E. & Grencis, R. K. An increase in epithelial cell apoptosis is associated with chronic intestinal nematode infection. Infect. Immun. 75, 1556–1564 (2007).
Gerbe, F. et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230 (2016).
Nadjsombati, M. S. et al. Detection of succinate by intestinal tuft cells triggers a Type 2 innate immune circuit. Immunity 49, 33–41.e7 (2018).
Schneider, C. et al. A metabolite-triggered tuft cell-ILC2 circuit drives small intestinal remodeling. Cell 174, 271–284 e14 (2018).
Nusse, Y. M. et al. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 559, 109–113 (2018).
Kotloff, K. L. et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209–222 (2013).
Checkley, W. et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect. Dis. 15, 85–94 (2015).
Platts-Mills, J. A. et al. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob. Health 3, e564–e575 (2015).
Khalil, I. A. et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob. Health 6, e758–e768 (2018).
Levy, J. et al. Clinical spectrum of X-linked hyper-IgM syndrome. J. Pediatr. 131, 47–54 (1997).
Gomez Morales, M. A. et al. Cryptosporidium parvum-specific CD4 Th1 cells from sensitized donors responding to both fractionated and recombinant antigenic proteins. Infect. Immun. 72, 1306–1310 (2004).
Hayward, A. R., Chmura, K. & Cosyns, M. Interferon-gamma is required for innate immunity to Cryptosporidium parvum in mice. J. Infect. Dis. 182, 1001–1004 (2000).
White, A. C. et al. Interferon-gamma expression in jejunal biopsies in experimental human cryptosporidiosis correlates with prior sensitization and control of oocyst excretion. J. Infect. Dis. 181, 701–709 (2000).
Leav, B. A. et al. An early intestinal mucosal source of gamma interferon is associated with resistance to and control of Cryptosporidium parvum infection in mice. Infect. Immun. 73, 8425–8428 (2005).
Tessema, T. S., Dauber, E. & Petry, F. Adoptive transfer of protective immunity from Cryptosporidium parvum-infected interferon-gamma and interleukin-12-deficient mice to naive recipients. Vaccine 27, 6575–6581 (2009).
Guerin, A. & Striepen, B. The biology of the intestinal intracellular parasite Cryptosporidium. Cell Host Microbe 28, 509–515 (2020).
Ehigiator, H. N., McNair, N. & Mead, J. R. Cryptosporidium parvum: the contribution of Th1-inducing pathways to the resolution of infection in mice. Exp. Parasitol. 115, 107–113 (2007).
Bedi, B., McNair, N. N. & Mead, J. R. Dendritic cells play a role in host susceptibility to Cryptosporidium parvum infection. Immunol. Lett. 158, 42–51 (2014).
Potiron, L. et al. Batf3-dependent intestinal dendritic cells play a critical role in the control of Cryptosporidium parvum Infection. J. Infect. Dis. 219, 925–935 (2019).
Barakat, F. M., McDonald, V., Di Santo, J. P. & Korbel, D. S. Roles for NK cells and an NK cell-independent source of intestinal gamma interferon for innate immunity to Cryptosporidium parvum infection. Infect. Immun. 77, 5044–5049 (2009).
Rohlman, V. C., Kuhls, T. L., Mosier, D. A., Crawford, D. L. & Greenfield, R. A. Cryptosporidium parvum infection after abrogation of natural killer cell activity in normal and severe combined immunodeficiency mice. J. Parasitol. 79, 295–297 (1993).
Bedi, B., McNair, N. N., Forster, I. & Mead, J. R. IL-18 cytokine levels modulate innate immune responses and cryptosporidiosis in mice. J. Eukaryot. Microbiol. 62, 44–50 (2015).
Choudhry, N., Petry, F., van Rooijen, N. & McDonald, V. A protective role for interleukin 18 in interferon gamma-mediated innate immunity to Cryptosporidium parvum that is independent of natural killer cells. J. Infect. Dis. 206, 117–124 (2012).
McDonald, V. et al. A potential role for interleukin-18 in inhibition of the development of Cryptosporidium parvum. Clin. Exp. Immunol. 145, 555–562 (2006).
McNair, N. N., Bedi, C., Shayakhmetov, D. M., Arrowood, M. J. & Mead, J. R. Inflammasome components caspase-1 and adaptor protein apoptosis-associated speck-like proteins are important in resistance to Cryptosporidium parvum. Microbes Infect. 20, 369–375 (2018).
Vinayak, S. et al. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523, 477–480 (2015).
Sateriale, A. et al. The intestinal parasite Cryptosporidium is controlled by an enterocyte intrinsic inflammasome that depends on NLRP6. Proc. Natl Acad. Sci. U.S.A. 118, e2007807118 (2021).
Ungar, B. L., Kao, T. C., Burris, J. A. & Finkelman, F. D. Cryptosporidium infection in an adult mouse model. Independent roles for IFN-gamma and CD4+ T lymphocytes in protective immunity. J. Immunol. 147, 1014–1022 (1991).
Harrington, L. E., Janowski, K. M., Oliver, J. R., Zajac, A. J. & Weaver, C. T. Memory CD4 T cells emerge from effector T-cell progenitors. Nature 452, 356–360 (2008).
Takeda, K. et al. Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity 8, 383–390 (1998).
Fuchs, A. et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity 38, 769–781 (2013).
Hunter, C. A. et al. Comparison of the effects of interleukin-1 alpha, interleukin-1 beta and interferon-gamma-inducing factor on the production of interferon-gamma by natural killer. Eur. J. Immunol. 27, 2787–2792 (1997).
Harrison, O. J. et al. Epithelial-derived IL-18 regulates Th17 cell differentiation and Foxp3(+) Treg cell function in the intestine. Mucosal Immunol. 8, 1226–1236 (2015).
Laurent, F. & Lacroix-Lamande, S. Innate immune responses play a key role in controlling infection of the intestinal epithelium by Cryptosporidium. Int J. Parasitol. 47, 711–721 (2017).
Rath, E., Moschetta, A. & Haller, D. Mitochondrial function—gatekeeper of intestinal epithelial cell homeostasis. Nat. Rev. Gastroenterol. Hepatol. 15, 497–516 (2018).
Moon, Y. Mucosal injuries due to ribosome-inactivating stress and the compensatory responses of the intestinal epithelial barrier. Toxins 3, 1263–1277 (2011).
Saeij, J. P. & Frickel, E. M. Exposing Toxoplasma gondii hiding inside the vacuole: a role for GBPs, autophagy and host cell death. Curr. Opin. Microbiol. 40, 72–80 (2017).
Yamamoto, M. et al. A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity 37, 302–313 (2012).
Liu, B. et al. Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G573–G584 (2013).
Maric-Biresev, J. et al. Loss of the interferon-gamma-inducible regulatory immunity-related GTPase (IRG), Irgm1, causes activation of effector IRG proteins on lysosomes, damaging lysosomal function and predicting the dramatic susceptibility of Irgm1-deficient mice to infection. BMC Biol. 14, 33 (2016).
Pollok, R. C., Farthing, M. J., Bajaj-Elliott, M., Sanderson, I. R. & McDonald, V. Interferon gamma induces enterocyte resistance against infection by the intracellular pathogen Cryptosporidium parvum. Gastroenterology 120, 99–107 (2001).
Kim, C. H., Hashimoto-Hill, S. & Kim, M. Migration and tissue tropism of innate lymphoid cells. Trends Immunol. 37, 68–79 (2016).
Gury-BenAri, M. et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231–1246 e13 (2016).
Park, E. et al. Toxoplasma gondii infection drives conversion of NK cells into ILC1-like cells. Elife. 8, e47605 (2019).
Weizman, O. E. et al. ILC1 confer early host protection at initial sites of viral infection. Cell 171, 795–808 e12 (2017).
Flanigan, T. et al. Cryptosporidium infection and CD4 counts. Ann. Intern. Med. 116, 840–842 (1992).
Navin, T. R. et al. Declining CD4+ T-lymphocyte counts are associated with increased risk of enteric parasitosis and chronic diarrhea: results of a 3-year longitudinal study. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20, 154–159 (1999).
O’Hara, S. P. et al. The human immunodeficiency virus type 1 tat protein enhances Cryptosporidium parvum-induced apoptosis in cholangiocytes via a Fas ligand-dependent mechanism. Infect. Immun. 75, 684–696 (2007).
Urban, J. F. Jr et al. IL-12 protects immunocompetent and immunodeficient neonatal mice against infection with Cryptosporidium parvum. J. Immunol. 156, 263–268 (1996).
Martinez-Lopez, M., Iborra, S., Conde-Garrosa, R. & Sancho, D. Batf3-dependent CD103+ dendritic cells are major producers of IL-12 that drive local Th1 immunity against Leishmania major infection in mice. Eur. J. Immunol. 45, 119–129 (2015).
Mashayekhi, M. et al. CD8alpha(+) dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity 35, 249–259 (2011).
Navabi, N. et al. Epithelial histone deacetylase 3 instructs intestinal immunity by coordinating local lymphocyte activation. Cell Rep. 19, 1165–1175 (2017).
Liu, Z. et al. Role of inflammasomes in host defense against Citrobacter rodentium infection. J. Biol. Chem. 287, 16955–16964 (2012).
Munoz, M. et al. Interleukin-22 induces interleukin-18 expression from epithelial cells during intestinal infection. Immunity 42, 321–331 (2015).
Jarret, A. et al. Enteric nervous system-derived IL-18 orchestrates mucosal barrier immunity. Cell 180, 50–63.e12 (2020).
Sellers, M. et al. Hirschsprung-associatedenterocolitis: observational study in a paediatric emergency care unit. An. Pediatr. 88, 329–334 (2018).
Teitelbaum, D. H., Caniano, D. A. & Qualman, S. J. The pathophysiology of Hirschsprung’s-associated enterocolitis: importance of histologic correlates. J. Pediatr. Surg. 24, 1271–1277 (1989).
Barakat, F. M., McDonald, V., Foster, G. R., Tovey, M. G. & Korbel, D. S. Cryptosporidium parvum infection rapidly induces a protective innate immune response involving type I interferon. J. Infect. Dis. 200, 1548–1555 (2009).
Ferguson, S. H. et al. Interferon-lambda3 promotes epithelial defense and barrier function against Cryptosporidium parvum infection. Cell. Mol. Gastroenterol. Hepatol. 8, 1–20 (2019).
Tretina, K., Park, E. S., Maminska, A. & MacMicking, J. D. Interferon-induced guanylate-binding proteins: Guardians of host defense in health and disease. J. Exp. Med. 216, 482–500 (2019).
Songhet, P. et al. Stromal IFN-gammaR-signaling modulates goblet cell function during Salmonella Typhimurium infection. PLoS ONE 6, e22459 (2011).
Monack, D. M., Bouley, D. M. & Falkow, S. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J. Exp. Med. 199, 231–241 (2004).
Lopez-Yglesias, A. H. et al. T-bet-dependent ILC1- and NK cell-derived IFN-gamma mediates cDC1-dependent host resistance against Toxoplasma gondii. PLoS Pathog. 17, e1008299 (2021).
Coers, J., Brown, H. M., Hwang, S. & Taylor, G. A. Partners in anti-crime: how interferon-inducible GTPases and autophagy proteins team up in cell-intrinsic host defense. Curr. Opin. Immunol. 54, 93–101 (2018).
Wilke G. et al. A stem-cell-derived platform enables complete Cryptosporidium development in vitro and genetic tractability. Cell Host Microbe. 26, 123–134 (2019).
Heo, I. et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 3, 814–823. (2018).
Dahan, S., Roth-Walter, F., Arnaboldi, P., Agarwal, S. & Mayer, L. Epithelia: lymphocyte interactions in the gut. Immunol. Rev. 215, 243–253 (2007).
Eberl, G. & Lochner, M. The development of intestinal lymphoid tissues at the interface of self and microbiota. Mucosal Immunol. 2, 478–485 (2009).
Zaph, C. et al. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature 446, 552–556 (2007).
Mamareli, P. et al. Epithelium-specific MyD88 signaling, but not DCs or macrophages, control acute intestinal infection with Clostridium difficile. Eur. J. Immunol. 49, 747–757 (2019).
Rauch, I. et al. NAIP-NLRC4 inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL-18 release via activation of caspase-1 and -8. Immunity 46, 649–659 (2017).
Chatterjee, M. & Ip, Y. T. Pathogenic stimulation of intestinal stem cell response in Drosophila. J. Cell Physiol. 220, 664–671 (2009).
Lee, K. Z. et al. Enterocyte purge and rapid recovery is a resilience reaction of the gut epithelium to pore-forming toxin attack. Cell Host Microbe 20, 716–730 (2016).
Ayyaz, A. & Jasper, H. Intestinal inflammation and stem cell homeostasis in aging Drosophila melanogaster. Front. Cell Infect. Microbiol. 3, 98 (2013).
Klover, P. J. et al. Loss of STAT1 from mouse mammary epithelium results in an increased Neu-induced tumor burden. Neoplasia 12, 899–905 (2010).
Hatton, R. D. et al. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity 25, 717–729 (2006).
Sateriale, A. et al. A Genetically tractable, natural mouse model of cryptosporidiosis offers insights into host protective immunity. Cell Host Microbe 26, 135–46 e5 (2019).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Mootha, V. K. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. U.S.A. 102, 15545–15550 (2005).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
This work was supported in part by funding from the Bill and Melinda Gates Foundation (OPP1183177) to B.S. and through grants from the U.S. National Institutes of Health to B.S. and C.A.H. (R01AI148249), R21 AI139895 to C.A.H., and fellowships and career awards to J.G. (T32AI055400), A.S. (K99AI137442), and A.G. (T32A1055400) and a Swiss National Science Foundation fellowship (191774) to S.B. We would also like to acknowledge the Commonwealth of Pennsylvania. This work was supported in part by funding from the Bill and Melinda Gates Foundation (OPP1183177) to B.S. and through grants from the U.S. National Institutes of Health to B.S. and C.A.H. (R01 AI148249), R21 AI139895 to C.A.H., and fellowships and career awards to J.G. (T32AI055400), A.S. (K99AI137442), and A.G. (T32A1055400) and a Swiss National Science Foundation fellowship (191774) to S.B. We would also like to acknowledge the Commonwealth of Pennsylvania.
Author information
Authors and Affiliations
Contributions
J.A.G., A.S., B.S. and C.A.H. conceived of the study; J.A.G. and A.S. conducted experiments. J.B.E. contributed pathologist expertise and histology scoring. AG provided the sequencing analysis, with technical support from D.P.B. S.S., Z.A.H. and L.M. contributed data acquisition. D.A.C., G.A.T. and M.Y. contributed mice and expertise. All authors approved the final version of this paper to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Gullicksrud, J.A., Sateriale, A., Engiles, J.B. et al. Enterocyte–innate lymphoid cell crosstalk drives early IFN-γ-mediated control of Cryptosporidium. Mucosal Immunol 15, 362–372 (2022). https://doi.org/10.1038/s41385-021-00468-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-021-00468-6
This article is cited by
-
Immunity to Cryptosporidium: insights into principles of enteric responses to infection
Nature Reviews Immunology (2024)
-
Evaluation of the therapeutic efficacy of some essential oils in experimentally immunosuppressed mice infected with Cryptosporidium parvum
Journal of Parasitic Diseases (2023)
-
The potential therapeutic effect of Nigella sativa and Zingiber officinale extracts versus Nitazoxanide drug against experimentally induced cryptosporidiosis in laboratory mice
Journal of Parasitic Diseases (2023)