Abstract
Memory CD4 T cells in tissues fulfill numerous functions that are critical for local immune homeostasis and protection against pathogens. Previous studies have highlighted the phenotypic and functional heterogeneity of circulating and tissue-resident memory CD4 T cells across different human tissues such as skin, lung, liver, and colon. Comparatively little is known in regard to memory CD4 T cells across tissues of the female reproductive tract (FRT). We examined CD4 T cells in donor-matched vaginal, ecto- and endocervical tissues, which differ in mucosal structure and exposure to external environmental stimuli. We hypothesized that this could be reflected by tissue-specific differences in the memory CD4 T cell compartment. We found differences in CD4 subset distribution across these tissues. Specifically, CD69+CD103+ CD4 T cells were significantly more abundant in vaginal than cervical tissues. In contrast, the transcriptional profiles of CD4 subsets were fairly conserved across FRT tissues. CD69+CD103+ CD4 T cells showed a TH17 bias independent of tissue niche. Our data suggest that FRT tissues affect T cell subset distribution but have limited effects on the transcriptome of each subset. We discuss the implications for barrier immunity in the FRT.
Similar content being viewed by others
Introduction
Memory T cells are crucial for rapidly mounting an effector immune response upon re-exposure to a pathogen.1 Different memory T cell subsets fulfill specific roles to accomplish immunosurveillance. While central memory T cells (TCM) maintain their potential to migrate to secondary lymphoid tissues, effector memory T cells (TEM) are characterized by the ability to migrate to peripheral tissues.2 In addition to these circulating memory subsets, there is also a subset of memory T cells that reside in the tissue for prolonged periods of time. These tissue-resident memory T cells (TRM) are activated in situ and possess a unique phenotype and gene expression profile.3,4,5,6 TRM were initially reported in the mouse model system,7 but are also present in human mucosal tissues,6,8,9,10,11,12,13 where they are thought to have a key role as protectors against pathogens.12,14,15 Although TRM cells are composed of both CD4 and CD8 T cell subsets, the majority of studies have focused on the protective role of CD8 TRM from pathogens mediated by direct cytotoxicity as well as IFNγ production. Importantly, the role of CD4 TRM is inherently more multi-faceted given that different CD4 T cell subsets carry out specific functions including protection against both intracellular and extracellular pathogens (TH1, TH2, TH17), immunoregulation (Treg), and tissue healing (TH17/22).12,16,17,18
CD4 TRM outnumber CD8 TRM in human mucosal barrier tissues such as the gut, lung, and oral mucosa.3,8,12,19 It has been shown that CD4 TRM display a large amount of diversity in phenotype depending on the tissue location.8,20 The composition of the T cell compartments is highly tissue-specific and CD4 TRM have been shown to produce differential cytokines depending on location.20,21
While the T cell compartment in human mucosal barrier tissues such as the lung and gut has been fairly well characterized, little is still known about the memory CD4 T cell compartment across tissues of the FRT. Vaginal and cervical tissues are adjacent mucosal tissues that differ significantly in their function and cellular structure. The upper FRT, including the endocervix (EndoCx), is comprised of type I mucosal surfaces covered by a single layer of columnar epithelial cells linked by tight junctions. The lower FRT, including the ectocervix (EctoCx) and vagina, are comprised of type II mucosal surfaces covered by multiple layers of non-keratinized stratified squamous epithelium.22,23 The upper FRT and lower FRT have distinct physiological functions and maintain different immune defense mechanisms—the lower FRT has a more acidic pH, local flora, and chemical signaling while the upper FRT is maintained sterilely.
We wanted to define how these anatomical and functional differences across endocervical, ectocervical, and vaginal mucosal tissues affect the memory CD4 T cell population in regard to subset distribution, cellular phenotype, and functional capacity. We used high parameter flow cytometry and RNAseq to examine the memory CD4 T cell compartment in paired blood and FRT tissues. While the general distribution of circulating vs. tissue-resident phenotype memory subsets appeared similar across tissues, we observed an enrichment of CD69+CD103+ double positive cells in the vaginal tissue (VT) when compared to the cervical tissues. We assessed the transcriptional and functional properties of this subset and found that CD69+CD103+ TRM expressed a transcriptome and effector cytokines consistent with TH17 responses. Importantly, the transcriptional profile of this and other CD4 memory subsets across FRT tissues appeared similar. Together, our data show that FRT tissues have a distinct T cell subset distribution but have limited effects on the transcriptome of each subset.
Results
Distinct CD4 to CD8 T cell ratios in FRT tissues
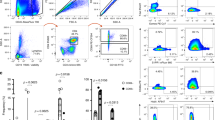
We first examined T cells in blood, VT, and non-malignant cervical tissue from healthy women undergoing elective hysterectomies or vaginal reconstructive surgeries for pelvic organ prolapse; prior to processing for single-cell suspensions, the cervix was dissected into EctoCx and EndoCx. This offered us the opportunity to compare T cells across adjacent mucosal sites in a donor-matched setup. We initially assessed the CD4 and CD8 T cell populations in the blood and these three tissue compartments (Fig. 1a, b). CD4 T cells were gated on CD25 expression to distinguish conventional CD4 T cells from regulatory T cells. We found that the CD4 to CD8 ratio was typically highest in the blood, but this trend was reversed in the vagina with generally a higher percentage of CD8 T cells (Fig. 1c). In addition to the significantly lower CD4:CD8 ratio in the VT, we also observed a lower CD4:CD8 ratio in the EctoCx but not the EndoCx when compared to the blood (Fig. 1c). As expected, the majority of CD4 T cells in the FRT had a CD45RA−CCR7− memory phenotype (Fig. 1d, e).
a Gating strategy for CD8 and CD4 T cells (top) and exclusion of CD25+CD127lo Treg from CD4 T cells (bottom) in blood, vagina (VT), ectocervix (EctoCx), and endocervix (EndoCx). Gated on Live, CD45+CD3+ cells. Numbers indicate percentages of the parent population. b Diagram of VT, EctoCx, and EndoCx within the female reproductive tract. c Ratio of CD4 to CD8 T cells. Lines connect samples from individual donors where all four compartments are represented. d Representative flow plots and e quantification of the memory (CCR7 and CD45RA) phenotype for CD4 Tconv cells. (Blood n = 9, VT n = 13, EctoCx n = 10, EndoCx n = 10) *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 generated by repeated measures one-way ANOVA with Tukey’s post test.
CD69+CD103+ cells are enriched in the vaginal tissue and express high levels of PD-1 and CCR5
CD69 and CD103 are widely used as biomarkers to identify human resident memory T (TRM) cells.3,15,24 It is important to note that not all TRM cells will express CD103 and that CD69 expression can also be a biomarker of recent activation. We found that a majority of CD4 T cells isolated from FRT were CD69 single positive and a substantial percentage of those also expressed CD103 (Fig. 2a, b). CD69+CD103+ CD4 T cells were absent from the blood, but present across all three tissue compartments. Notably, the fraction of CD69+CD103+ CD4 T cells was significantly higher in the VT than either the EndoCx or EctoCx (Fig. 2c).
a Representative flow plots for tissue memory (CD69 and CD103) phenotype of CD4 Tconv cells. Gated on Live, CD45+, CD3+, CD4+, CD25−CD127+ cells, b proportions of CD69+CD103− (purple), CD69+CD103+ (blue), CD69−CD103+ (orange), and CD69−CD103− (green), and c percent of CD69+CD103+ on CD4 Tconv cells in blood n = 8, vagina (VT) n = 14, ectocervix (EctoCx) n = 9, and endocervix (EndoCx) n = 9. Lines connect samples from individual donors where all four compartments are represented. d Representative flow plots and e quantification of PD-1 positive CD4 Tconv cells in blood and mucosal tissues. Gated on Live, CD45+, CD3+, CD4+, CD25−CD127+ cells. f Percent PD-1+CD4+ T cell in each memory population subset. Lines connect samples from individual donors where all four compartments are represented. g Representative flow plots and h quantification of CCR5 positive CD4 Tconv cells in blood and mucosal tissues. Gated on Live, CD45+, CD3+, CD4+, CD25−CD127+ cells. i Percent CCR5+CD4+ T cell in each memory population subset. Lines connect samples from individual donors where all four compartments are represented. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 generated by repeated measures one-way ANOVA with Tukey’s post test.
We next assessed PD-1 expression as an additional biomarker for TRM cells. PD-1 is highly expressed on effector T cells and exhausted/dysfunctional T cells, but also expressed on effector memory T cells in the blood25 and TRM in tissues.6 Tissue CD4 T cell PD-1 expression was predominantly limited to CD69+ cells, whereas a small proportion of CD4 T cells in the blood expressed PD-1, but primarlily in the CD69− fraction (Fig. 2d). Overall, PD-1 expression was increased in CD4 T cells from each of the three genital tract mucosal tissue types compared to CD4 T cells from the blood (Fig. 2e). Furthermore, PD-1 expressing cells were enriched in the CD69+CD103+ population in the vagina and EctoCx compared to all other cell subsets (Fig. 2f). Thus, in line with previous reports of T cells in other tissues, CD4 T cells within the FRT also highly co-express key markers associated with the TRM phenotype.6
We further characterized the phenotype of all memory CD4 T cells in the vaginal and cervical tissues. We and others have previously shown that CCR5+ CD4 T cells are part of healthy barrier immunity12,26 and are stably maintained as part of the CD4 tissue-resident memory population in healthy as well as inflamed tissue.12 To determine if this is also true across tissues in the FRT, we measured CCR5 expression on CD4 T cells by flow cytometry. Over half of the CD4 T cells in the FRT express CCR5 across all three tissue sites (Fig. 2g, h). We next examined CCR5 expression on CD4 T cells with different CD69 and CD103 expression patterns and we found that CCR5+ cells were enriched in the CD69+CD103+ tissue-resident memory T cell population (Fig. 2i). The CD69−CD103− population had the lowest proportion of CCR5+ cells, while the CD69+CD103+ population almost uniformly expressed CCR5 (Fig. 2i). All three tissues showed similar patterns, although the differences were most striking in the EndoCx.
CD69+CD103+ CD4 T cells sorted from vaginal tissue have a unique transcriptional profile
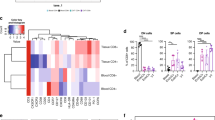
Given that we saw an enrichment of CD69+CD103+ CD4 T cells in the vagina, we next wanted to determine if this subset has unique functional features compared to the other tissue CD4 T cell subsets. To assess the transcriptional and functional potential of these populations, we performed bulk RNA sequencing (RNAseq) as previously described27 (Fig. 3a). From four donors, CD4 T cells were sorted based on CD69−CD103−, CD69+CD103−, or CD69+CD103+ expression from donor-matched VT, EctoCx, and EndoCx tissues. From the donor-matched blood just CD69−CD103− and CD69+CD103− expressing subsets were sorted as CD69+CD103+ cells were not present in appreciable numbers in the circulation. We first examined the transcriptional overlap and changes between CD69/CD103 CD4 T cell subsets within VT to determine if these subsets could be differentiated by a unique transcriptional profile. To determine transcriptome differences, we examined differentially expressed genes (DEGs) using an absolute log2-fold change cutoff of 1 and an FDR cutoff of 5%. We first assessed the number of DEG between the different CD4 subsets within the VT compartment (Fig. 3b). The largest number of DEGs were found between the CD69−CD103− and CD69+CD103+ populations of CD4 T cells (over 200 DEG combined) (Fig. 3b). In general, the two CD69+ expressing subsets expressed largely overlapping DEGs but had pronounced differences compared to the CD69− subset (Fig. 3c). Finally, we observed transcriptional differences that indicated a potential divergence in function in these subsets (Fig. 3d). Of note, RORC and other IL-17-associated genes appeared highly expressed in the CD69+CD103+ subset (Fig. 3d).
a Schematic diagram showing experimental setup for RNAseq experiment and representative flow plots and RNAseq sort gates for CD69−CD103− (green), CD69+CD103− (purple), and CD69+CD103+ (blue) CD4 T cells. Cells were gated on Live, CD45+CD3+CD4+CCR7− (n = 4 donors). b The number of DEGs between tissue memory CD4 T cell comparisons in the vagina. c The percentage of overlapping DEGs represented by circles for each cell subset comparison in the vagina. Percentages were calculated by dividing the number of shared DEGs between two cell subsets by the total number of DEGs. Larger circles represent more overlapping genes and smaller circles represent fewer overlapping genes. d Heatmap of DEGs from cell populations sorted from the vagina. An absolute log2-fold change cutoff of 1 and an FDR cutoff of 5% were used to determine DEGs. Only the top DEGs from each comparison are shown.
CD69+CD103+ CD4 TRM cells in VT are enriched for a TH17 gene signature
Given the difference in RORC expression in the CD69+CD103+ subset compared to the other subsets in the VT, we wanted to further examine the transcriptional changes and determine if there was evidence of a TH17 gene signature in the CD69+CD103+ CD4 T cells. We used a previously published TH17 gene signature12 to determine if there was an enhanced TH17 signature in the CD4 TRM-like cells in the VT (Fig. 4a). When we looked at the TH17 gene set enrichment by barcode plot, we found that the TH17-associated genes were upregulated in the CD69+CD103+ T cell compartment of the VT compared to the CD69+CD103− CD4 T cell subset (Fig. 4b). We further confirmed this finding by looking at individual normalized gene expression from the sorted CD4 T cell populations. In the blood, gene transcript levels for TH17-related genes were not enriched in any particular cell subset (Fig. 4c). In the VT however, the TH17 gene transcript levels were increased in CD69+CD103+ CD4 T cells compared to the other subsets (Fig. 4d). Overall, these data indicate that the CD69+CD103+ CD4 TRM-like cell subset is more prone to express IL-17-related genes compared to the other subsets.
a List of TH17 associated genes used for gene set enrichment analysis. b Gene set enrichment analysis showing TH17 associated genes upregulated (red) or downregulated (blue) in CD69+CD103+ cells isolated from the vagina compared to CD69+CD103− CD4 T cells. c Boxplot graph showing normalized gene expression levels of TH17 associated genes in CD69−CD103− (green) and CD69+CD103− (purple) in the blood. d Boxplot graph of normalized gene expression levels of Th17 associated genes in CD69−CD103− (green), CD69+CD103− (purple), and CD69+CD103+ (blue) from the vagina. c, d Each dot represents cells sorted from one individual (n = 4).
IL-17 in the VT is primarily produced by CD69+CD103+ CD4 TRM
We next wanted to confirm if the transcript-based findings translated into increased cytokine production and a functional TH17 profile. Functional ex vivo assays were only technically possible when a sufficient number of T cells could be isolated from the obtained tissues. In these instances, CD4 T cells from VT were incubated with phorbol 12-myristate 13-acetate (PMA)/ionomycin, followed by assessment of cytokine production and expression of CD69 and CD103 by flow cytometry (Fig. 5). We observed that the CD4 T cells from the VT were capable of producing IL-17A upon restimulation (Fig. 5a, b). We next wanted to determine whether CD69/CD103 expressing subsets were preferentially able to produce this cytokine. CD69−CD103+ CD4 T cells were rare in VT and so cytokine production could not be accurately assessed (Fig. 5a, b). We found no to very limited IL-17A expression by CD69−CD103− cells, whereas a sizeable fraction of CD69+CD103+ cells and, to a lesser extent, CD69+CD103− cells produced IL-17A after stimulation (Fig. 5a–c). When we compared cytokine production by CD4 T cells from the VT compared to donor-matched blood, CD4 T cells from the VT produced more IL-17A after stimulation (Fig. 5d). We also examined expression of IL-2, TNFα, and IFNγ in CD4 T cells from blood and FRT (Supplementary Fig. 1A–C). Overall, we observed similar expression of TNFα, and IFNγ in CD4 T cells regardless of tissue origin, while IL-2 expression was significantly decreased in CD4 T cells isolated from the FRT. Subsetting of the CD4 T cells from the FRT based on CD69 and CD103 expression revealed comparable IL-2, TNFα, and IFNγ expression between CD69+CD103− and CD69+CD103+ CD4 T cell populations, while CD69−CD103− CD4 T cells were overall less responsive to restimulation (Supplementary Fig. 1A). In sum, CD4 T cells in the VT were capable of producing IL-17A, and CD69+CD103+ TRM-like CD4 T cells in the VT produced the majority of the IL-17A compared to CD69−CD103− or CD69+CD103− CD4 T cell subsets.
CD4 Tconv cells isolated from vaginal tissue were stimulated with PMA/Ionomycin (or left unstimulated), stained for tissue memory markers and IL-17A production, and analyzed by flow cytometry. A sufficient number of T cells to undergo stimulation was isolated from tissues in three individuals, but only two individuals had enough cells to adequately examine CD69 and CD103 expressing populations. Representative flow plots (a, b) and quantification (c) from two individuals looking at percent IL-17A positive of CD4 Tconv in tissue memory subsets CD69−CD103− (green), CD69+CD103− (purple), and CD69+CD103+ (blue). d Cells were gated on lymphocytes, then live, CD45+, CD3+, and CD4+ T cells, then gated on the CD45RA−CCR7+, CD45RA−CCR7−, and CD45RA+CCR7− T cell subsets, excluding CD45RA+CCR7+ naïve cells from analysis. Percent IL-17A of CD4+ Tconv cells in the blood compared to the vagina (n = 3). *p ≤ 0.05 generated by two-tailed paired t-test.
CD69+CD103+ CD4 TRM TH17 gene enrichment is conserved across adjacent mucosal tissues
We next wanted to determine if the TH17 gene signature found in VT was conserved across CD69+CD103+ CD4 T cells in all three FRT sites. We therefore compared the same TH17 gene set (Fig. 4a) across the sorted cell populations from the cervical tissues (Fig. 3a). The EctoCx was very similar to the VT in that CD69+CD103+ CD4 T cells were highly enriched for a TH17 signature compared to the CD69+CD103− CD4 population (Fig. 6a). When comparing the individual TH17 gene transcripts between cell subsets in the EctoCx, CD69+CD103+ CD4 T cells had the highest transcript levels for all of the TH17-related genes assessed (Fig. 6b). The CD69+CD103+ CD4 T cells sorted from the EndoCx had a weaker, albeit positive, association with the TH17 gene signature (Fig. 6c). We also found that the CD69+CD103− subset had comparable levels of TH17-related gene expression to the CD69+CD103+ population in the EndoCx (Fig. 6d). Together, these data indicate that the CD69+CD103+ CD4 T cell subset is associated with a TH17 signature across all FRT tissues.
a Gene set enrichment analysis showing TH17 associated genes upregulated (red) or downregulated (blue) in CD69+CD103+ compared to CD69+CD103− cells isolated from the ectocervix (EctoCx). b Boxplot graph showing normalized gene expression levels of TH17 associated genes in CD69−CD103− (green), CD69+CD103− (purple), and CD69+CD103+ (blue) in the EctoCx. c Gene set enrichment analysis showing TH17 associated genes upregulated (red) or downregulated (blue) in CD69+CD103+ compared to CD69+CD103− cells isolated from the endocervix (EndoCx). d Boxplot graph of normalized gene expression levels of TH17 associated genes in CD69−CD103− (green), CD69+CD103− (purple), and CD69+CD103+ (blue) from the EndoCx. Each dot represents cells sorted from one individual (n = 4).
Tissue memory CD4 T cell subsets in adjacent mucosal sites have largely shared transcriptional profiles
Finally, given the transcriptional congruence of CD69+CD103+ CD4 T cells in the VT, we also wanted to assess CD69+CD103− and CD69−CD103− CD4 T cell subsets in a similar manner across FRT tissues. We examined the RNAseq data obtained from each CD4 T cell subset (CD69−CD103−, CD69+CD103−, and CD69+CD103+) across the three FRT tissue locations (Fig. 7a). There were very few DEG between CD69−CD103− tissue CD4 T cells from the vagina, EctoCx, and EndoCx. Similarly, few genes were differentially expressed by CD69+CD103− or CD69+CD103+ CD4 T cells when comparing between FRT sites, although there were more DEG for these populations between the VT and either cervical tissue (Fig. 7b and Supplementary Fig. 2). Overall, our data suggest that the functional properties of CD4 T cell subsets across these three FRT are rather conserved.
Discussion
Mouse model studies have provided compelling evidence that the functional properties of CD4 T cells can be modulated by the tissue in which the T cells reside.18,28 Determining if such tissue-mediated adaptation of T cells also occurs in human tissues is challenging, but has been done by Farber et al. using tissues from recently deceased organ donors.6,8 Some studies have been able to examine the T cell compartment across several different tissues in a donor-matched manner in the FRT,13,29,30,31 yet the immune cell compartment in human FRT has remained poorly characterized despite the critical role of the FRT in regards to barrier immunity as well as reproductive health. To compare the CD4 T cell compartment across FRT tissues, we collected vaginal and cervical tissues following elective hysterectomies to analyze CD4 memory T cell distribution in donor-matched vaginal, ectocervical, and endocervical tissues. A donor-matched analysis of tissues and blood is ideal to identify the extent to which tissue structure, function, and environment shape the composition and functional properties of the T cell compartment.
We used a unique combination of flow cytometry, RNAseq and ex vivo functional assays to define differences in the T cell compartment of these FRT tissues. We specifically wanted to focus on the tissue-resident CD4 T cell compartment given their pleiotropic roles in tissues ranging from protection to maintaining tissue homeostasis. We first examined the CD4 to CD8 T cell subset distribution in all tissues. We found that CD4 T cells were more abundant than CD8 T cells in the EndoCx and EctoCx. This is akin to other mucosal sites such as the lung, gut, and oral mucosa, which also have a bigger memory CD4 T cell than CD8 T cell compartment.3,12,19 Interestingly, we found that the vaginal tract had nearly a 1:1 CD4:CD8 T cell ratio. An increase in CD8 T cell numbers could reflect the need for heightened antiviral and antibacterial immune surveillance in the VT compared to cervical tissues. The mechanism driving this increase in CD8 T cells is unknown, but it is noteworthy that activated tissue-resident memory CD8 T cells can recruit peripheral memory CD8 T cells in an IFNγ-dependent manner in a mouse model system.32 Of note, another mouse model study indicated that the VT environment may restrict the immune response to RNA viruses.33 Whether the increase in CD8 T cells is an attempt to compensate for this dampened antiviral innate immune response remains unclear and additional studies will be required to determine the biological significance of this observation.
We further examined the memory CD4 T cell compartment and found that its composition differs in the VT compared to cervical tissue. In line with previous reports we used the biomarkers CD69 and CD103 to indicate tissue-residence of T cells. CD69+CD103+ CD4 TRM cells were more abundant in the CD4 compartment of the VT compared to the cervical tissues. Using RNAseq, we determined that these CD69+CD103+ CD4 TRM cells from the VT were characterized by enhanced IL-17A production at a transcriptional and protein level compared to the CD69−CD103− population. Similarly to the even distribution of CD4 to CD8 T cells in the VT, this again points toward strengthened layers of immune protection in the VT. IL-17 can exert antibacterial function and is as such a key cytokine for barrier immunity.34 The vagina in particular is continuously exposed to the vaginal microbiota, which is predominantly composed of Lactobacillus species, but can become dysregulated and lead to bacterial vaginosis.35 Genital concentrations of IL-17 were found to be increased in women with bacterial sexually transmitted infections,36 suggesting that some microbial exposures can promote TH17 responses. TH17 cells clearly can have pro-inflammatory functions, but also have been shown to help maintain mucosal barrier integrity by promoting the formation of tight junctions between intestinal epithelial cells.37 Therefore, these cells may play an important role during homeostasis as well as when mounting a response in the context of a pathogen in the FRT. The increased propensity for inflammation and exposure to microbial flora in the vagina could also enhance cytokines known to play a role in survival and retention of TRM, including IL-15, IFNγ, and TGFβ.38 Overall, our data suggest that differences in primary physiologic function at least in part dictate the requirements for T cell-mediated immunity. We also considered that differences in tissue organization and structure between VT and the cervix could be reflected by changes in the transcriptional profile of T cells. However, the transcriptome of all three CD4 subsets was largely conserved across donor-matched mucosal FRT sites despite these differences in tissue organization and structure.
Although our study was not designed to assess differences in sex hormones across donors and tissues, we also consider that such differences could contribute to changes in the T cell compartment. Sex hormones have been shown to affect immune cells; in particular, endogenous progesterone, which is produced locally in the ovary by the corpus luteum, has an anti-inflammatory effect.39 In vitro studies have demonstrated that endogenous progesterone suppresses activation of antigen presenting cells40,41,42 and increases secretion of the anti-inflammatory cytokine IL-10 by DCs. Endogenous progesterone has also been shown to skew T cell responses toward a TH2 type response43,44 and promote the generation of Treg cells.45,46 Thus, it is likely that progesterone plays a role in modulating tissue T cell responses in the FRT. Although it is not currently appreciated, it would be important to investigate if there are differences in progesterone and estrogen concentrations and/or gradients across tissues of the FRT, which could impact the local immune microenvironment and contribute to the phenotypic and numerical differences in tissue T cell subsets we observed. We predict that gradients of local progesterone could play a role in a tolerance gradient within the FRT, wherein it functions to assist in maintaining immune quiescence in the upper FRT—most critically the uterus, to allow for implantation and promote fetal tolerance—while still allowing for active immune surveillance in the lower FRT, where more microbial exposures are likely to occur. Further studies are required to investigate this role of local sex hormones on tissue T cell phenotype and function in the FRT, including investigations on the concentrations of endogenous progesterone within the different tissue types and the effect of exposure to progesterone on tissue T cells. The study of immune activation mediated by CD4 T cells in the FRT has implications for both fertility and pregnancy as well as for sexually transmitted infections, including HIV, as activated CD4 T cells have the potential to be target cells for the latter.
In summary, we demonstrate that CD8 T cells and the TH17-biased CD69+ CD103+ CD4 T cell subset were more abundant in vaginal than cervical tissues, likely reflecting the VT’s physiologic function as a critical barrier tissue. Importantly, the transcriptional profiles of different CD4 T cell subsets were fairly conserved across FRT tissues indicating that these tissues affect T cell subset distribution but have limited effects on the transcriptome of each subset. Our data provide a characterization of the CD4 memory landscape across vaginal, endocervical, and ectocervical tissues and as such provide a T cell reference dataset to further study FRT tissues in the context of different pathologies or immune interventions.
Materials and methods
Human samples
Human blood, cervical, and VT were collected from healthy women undergoing elective hysterectomy. Additional VT were collected from women undergoing vaginal reconstructive surgeries for pelvic organ prolapse. Any samples suspected of malignancy were excluded from collection. All participants signed a written informed consent prior to inclusion in the study and the protocols were approved by the institutional review board at the Fred Hutchinson Cancer Research Center and the University of Washington (4323).
Tissue processing
Blood was collected into ACD tubes and mixed. Blood was diluted 1:2 in PBS, layered on a Lymphoprep (StemCell Technologies) gradient and centrifuged at 1200 rpm for 20 min. The PBMC layer was then removed and washed with PBS. Tissues were transported from the clinic in ice cold PBS and immediately processed upon arrival (within 1–2 h of removal). Surgical samples were trimmed to 2 mm pieces, and digested with collagenase II (700 units/ml, Sigma-Aldrich), and DNAse I (1 unit/ml, Sigma-Aldrich) for two subsequent 30 min digestions at 37 °C as previously described.47
Flow cytometry and cell sorting
Single-cell suspensions were washed with PBS and stained with LIVE/DEAD Fixable Dead Cell Stain (Thermo Fisher) for 15 min, followed by staining with an optimized antibody cocktail for 20 min at room temperature. Cells were then washed with FACS buffer (PBS containing 2% FBS) and fixed in PBS containing 1% paraformaldehyde (Sigma-Aldrich). For samples requiring intracellular staining, cells were processed using the Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher) according to the manufacture’s instruction. Samples were acquired using a FACSymphony (BD Biosciences). Data were analyzed with FlowJo software (version 9.8.8 or higher, BD Biosciences). All sorting experiments were carried out on freshly isolated cells using a FACSAriaII (BD Biosciences). Antibodies used include: CD45 (HI30), CD103 (Ber-ACT8), CD45RA (HI100, Biolegend), CD3 (UCHT1), CD69 (FN50), CD8 (SK1 or RPA-T8), CCR7 (G043H7, Biolegend), CD25 (M-A251), CD127 (HIL-7R-M21), CD4 (RPA-T4), CCR5 (2D7), PD-1 (EH12.1, Biolegend), Granzyme B (GB11), Foxp3 (259D/C7), CTLA4 (BNI3), IL-17A (N49-653), IL-2 (5344.111), (TNFα (Mab11), IFNγ (B27). All antibodies were from BD Biosciences unless otherwise noted.
Re-stimulation assay
T cells were quantified using a Guava easyCyte (Millipore). Single-cell suspensions from blood and tissue were put in complete media at a concentration of 2 × 104 T cells per condition (consistent between blood and tissue for each donor). Cells were left untreated or stimulated with 1 ng/ml PMA and 0.1 μM ionomycin for 5 h at 37 °C. All stimulations were performed in the presence of Brefeldin A (Golgi plug, BD Biosciences). Following stimulation, cells were stained as described above for flow cytometry.
RNAseq library generation
We performed RNAseq on 250 sorted cells for each sample from blood and tissue as previously described.27 In total, 44 samples were sequenced from n = 4 individuals. Samples were as follows (Live, CD45+CD3+CD4+): blood CD103−CD69−, blood CD103−CD69+, vaginal CD103−CD69−, vaginal CD103−CD69+, vaginal CD103+CD69+, EctoCx CD103−CD69−, EctoCx CD103−CD69+, EctoCx CD103+CD69+, EndoCx CD103−CD69−, EndoCx CD103−CD69+, EndoCx CD103+CD69+. Briefly, cells were sorted directly into SMARTer® v4 lysis reagents (Clontech). Cells were lysed and cDNA was synthesized. After amplification, sequencing libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina). Barcoded single-cell libraries were pooled and quantified using a Qubit® Fluorometer (Life Technologies).
RNAseq sequencing and alignment of libraries
Single-read sequencing of the libraries was carried out on a HiSeq2500 sequencer (Illumina) with 58-base pair reads, using TruSeq v4 Cluster and SBS kits (Illumina) with a target depth of five million reads. RNAseq data were aligned to the human genome (UCSC Human Genome Assembly, reference sequence GRCh38) using STAR (v2.4.2a),48 and gene expression quantification was performed using RSEM (v1.2.22).49 Genes with <13 nonzero read counts were filtered out, leaving 14,720 expressed genes for downstream analysis. Samples with <500,000 reads; 7000 detected genes; and an exon rate <50% were discarded. All libraries except one passed these quality criteria, ending up with 43 samples.
RNAseq statistical analysis
Raw count data were imported into R (v3.6.2). The edgeR Bioconductor package was used to calculate normalization factors to scale the raw library sizes,50 followed by a voom normalization from the limma Bioconductor package.51,52 It transforms count data to log2 counts per million and estimates the mean-variance relationship to compute appropriate observation-level weights.
Differential expression analyses were performed using the limma statistical framework and associated R package.52,53 A linear model was fitted to each gene, and empirical Bayes moderated t-statistics (two-tailed) were used to assess differences in expression. Contrasts comparing tissues and/or cell populations were tested. Intraclass correlations were estimated using the duplicate correlation function of the limma package to account for measures originating from the same donors.54 An absolute log2-fold change cutoff of 1 and an FDR cutoff of 5% were used to determine DEGs.
Gene set enrichment analysis was performed using the R function Camera implemented within the limma R package.55 The same contrasts as above were investigated. A FDR cutoff of 5% was used to determine significant gene sets. KEGG (v6), BioCarta (v6), and curated pathways were used as gene sets. Those gene sets were downloaded from MSigDB database.56
Statistical analyses of cell frequencies
For flow cytometry data, a paired t-test, linear regression or repeated measures one-way ANOVA with Tukey’s post test was performed as described in figure legends. Two-sided p values >0.05 were considered not significant, and values denoted with (*) symbols reflect significance levels as follows: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001.
Data availability
RNAseq data available as GEO number GSE163260.
References
Williams, M. A. & Bevan, M. J. Effector and memory CTL differentiation. Annu. Rev. Immunol. 25, 171–192 (2007).
Sallusto, F., Lenig, D., Forster, R., Lipp, M. & Lanzavecchia, A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999).
Sathaliyawala, T. et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity 38, 187–197 (2013).
Ariotti, S., Haanen, J. B. & Schumacher, T. N. Behavior and function of tissue-resident memory T cells. Adv. Immunol. 114, 203–216 (2012).
Milner, J. J. & Goldrath, A. W. Transcriptional programming of tissue-resident memory CD8(+) T cells. Curr. Opin. Immunol. 51, 162–169 (2018).
Kumar, B. V. et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20, 2921–2934 (2017).
Wakim, L. M., Waithman, J., van Rooijen, N., Heath, W. R. & Carbone, F. R. Dendritic cell-induced memory T cell activation in nonlymphoid tissues. Science 319, 198–202 (2008).
Thome, J. J. et al. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 159, 814–828 (2014).
Cheuk, S. et al. CD49a expression defines tissue-resident CD8(+) T cells poised for cytotoxic function in human skin. Immunity 46, 287–300 (2017).
Pallett, L. J. et al. IL-2(high) tissue-resident T cells in the human liver: sentinels for hepatotropic infection. J. Exp. Med. 214, 1567–1580 (2017).
Shin, H. & Iwasaki, A. Tissue-resident memory T cells. Immunol. Rev. 255, 165–181 (2013).
Woodward Davis, A. S. et al. The human tissue-resident CCR5(+) T cell compartment maintains protective and functional properties during inflammation. Sci. Transl. Med. 11 https://doi.org/10.1126/scitranslmed.aaw8718 (2019).
Pattacini, L. et al. A pro-inflammatory CD8+ T-cell subset patrols the cervicovaginal tract. Mucosal Immunol. 12, 1118–1129 (2019).
Zhu, J. et al. Immune surveillance by CD8alphaalpha+ skin-resident T cells in human herpes virus infection. Nature 497, 494–497 (2013).
Clark, R. A. et al. Skin effector memory T cells do not recirculate and provide immune protection in alemtuzumab-treated CTCL patients. Sci. Transl. Med. 4, 117ra117 (2012).
Turner, D. L. & Farber, D. L. Mucosal resident memory CD4 T cells in protection and immunopathology. Front. Immunol. 5, 331 (2014).
Schreiner, D. & King, C. G. CD4+ memory T cells at home in the tissue: mechanisms for health and disease. Front. Immunol. 9, 2394 (2018).
Nguyen, Q. P., Deng, T. Z., Witherden, D. A. & Goldrath, A. W. Origins of CD4(+) circulating and tissue-resident memory T-cells. Immunology 157, 3–12 (2019).
Turner, D. L. et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 7, 501–510 (2014).
Wong, M. T. et al. A high-dimensional Atlas of human T cell diversity reveals tissue-specific trafficking and cytokine signatures. Immunity 45, 442–456 (2016).
Szabo, P. A., Miron, M. & Farber, D. L. Location, location, location: tissue resident memory T cells in mice and humans. Sci. Immunol. 4 https://doi.org/10.1126/sciimmunol.aas9673 (2019).
Iwasaki, A. Antiviral immune responses in the genital tract: clues for vaccines. Nat. Rev. Immunol. 10, 699–711 (2010).
Kumamoto, Y. & Iwasaki, A. Unique features of antiviral immune system of the vaginal mucosa. Curr. Opin. Immunol. 24, 411–416 (2012).
Kumar, B. V., Connors, T. J. & Farber, D. L. Human T cell development, localization, and function throughout life. Immunity 48, 202–213 (2018).
Duraiswamy, J. et al. Phenotype, function, and gene expression profiles of programmed death-1(hi) CD8 T cells in healthy human adults. J. Immunol. 186, 4200–4212 (2011).
Hladik, F., Lentz, G., Delpit, E., McElroy, A. & McElrath, M. J. Coexpression of CCR5 and IL-2 in human genital but not blood T cells: implications for the ontogeny of the CCR5+ Th1 phenotype. J. Immunol. 163, 2306–2313 (1999).
Voillet, V. et al. Human MAIT cells exit peripheral tissues and recirculate via lymph in steady state conditions. JCI Insight 3 https://doi.org/10.1172/jci.insight.98487 (2018).
Cipolletta, D., Kolodin, D., Benoist, C. & Mathis, D. Tissular T(regs): a unique population of adipose-tissue-resident Foxp3+CD4+ T cells that impacts organismal metabolism. Semin. Immunol. 23, 431–437 (2011).
Givan, A. L. et al. Flow cytometric analysis of leukocytes in the human female reproductive tract: comparison of fallopian tube, uterus, cervix, and vagina. Am. J. Reprod. Immunol. 38, 350–359 (1997).
Rodriguez-Garcia, M., Barr, F. D., Crist, S. G., Fahey, J. V. & Wira, C. R. Phenotype and susceptibility to HIV infection of CD4+ Th17 cells in the human female reproductive tract. Mucosal Immunol. 7, 1375–1385 (2014).
Trifonova, R. T., Lieberman, J. & van Baarle, D. Distribution of immune cells in the human cervix and implications for HIV transmission. Am. J. Reprod. Immunol. 71, 252–264 (2014).
Schenkel, J. M., Fraser, K. A., Vezys, V. & Masopust, D. Sensing and alarm function of resident memory CD8(+) T cells. Nat. Immunol. 14, 509–513 (2013).
Khan, S. et al. Dampened antiviral immunity to intravaginal exposure to RNA viral pathogens allows enhanced viral replication. J. Exp. Med. 213, 2913–2929 (2016).
McGeachy, M. J., Cua, D. J. & Gaffen, S. L. The IL-17 family of cytokines in health and disease. Immunity 50, 892–906 (2019).
Ma, B., Forney, L. J. & Ravel, J. Vaginal microbiome: rethinking health and disease. Annu. Rev. Microbiol. 66, 371–389 (2012).
Masson, L. et al. Relationship between female genital tract infections, mucosal interleukin-17 production and local T helper type 17 cells. Immunology 146, 557–567 (2015).
Kinugasa, T., Sakaguchi, T., Gu, X. & Reinecker, H. C. Claudins regulate the intestinal barrier in response to immune mediators. Gastroenterology 118, 1001–1011 (2000).
Iijima, N. & Iwasaki, A. Tissue instruction for migration and retention of TRM cells. Trends Immunol. 36, 556–564 (2015).
Hall, O. J. & Klein, S. L. Progesterone-based compounds affect immune responses and susceptibility to infections at diverse mucosal sites. Mucosal Immunol. 10, 1097–1107 (2017).
Jones, L. A. et al. Differential modulation of TLR3- and TLR4-mediated dendritic cell maturation and function by progesterone. J. Immunol. 185, 4525–4534 (2010).
Jones, L. A. et al. Toll-like receptor-4-mediated macrophage activation is differentially regulated by progesterone via the glucocorticoid and progesterone receptors. Immunology 125, 59–69 (2008).
Butts, C. L. et al. Progesterone regulation of uterine dendritic cell function in rodents is dependent on the stage of estrous cycle. Mucosal Immunol. 3, 496–505 (2010).
Miyaura, H. & Iwata, M. Direct and indirect inhibition of Th1 development by progesterone and glucocorticoids. J. Immunol. 168, 1087–1094 (2002).
Piccinni, M. P. et al. Progesterone favors the development of human T helper cells producing Th2-type cytokines and promotes both IL-4 production and membrane CD30 expression in established Th1 cell clones. J. Immunol. 155, 128–133 (1995).
Lee, J. H., Lydon, J. P. & Kim, C. H. Progesterone suppresses the mTOR pathway and promotes generation of induced regulatory T cells with increased stability. Eur. J. Immunol. 42, 2683–2696 (2012).
Lee, J. H., Ulrich, B., Cho, J., Park, J. & Kim, C. H. Progesterone promotes differentiation of human cord blood fetal T cells into T regulatory cells but suppresses their differentiation into Th17 cells. J. Immunol. 187, 1778–1787 (2011).
McKinnon, L. R. et al. Optimizing viable leukocyte sampling from the female genital tract for clinical trials: an international multi-site study. PLoS ONE 9, 1–11 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Zafirova, B., Wensveen, F. M., Gulin, M. & Polic, B. Regulation of immune cell function and differentiation by the NKG2D receptor. Cell. Mol. life Sci. 68, 3519–3529 (2011).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, 1–17 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 https://doi.org/10.2202/1544-6115.1027 (2004).
Smyth, G. K., Michaud, J. & Scott, H. S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21, 2067–2075 (2005).
Wu, D. & Smyth, G. K. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 40, e133 (2012).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Acknowledgements
We thank Veronica Davé for her input on statistical analysis and members of the Lund and Prlic labs for critical discussions. This work was supported by NIH grants DP2 DE023321 (M.P.), T32AI07140 (A.S.W.D.), R01 AI141435 and AI131914 (J.M.L.), and R01 DA040386 (F.H.). A.S.W.D. was a Doug and Maggie Walker Fellow.
Author information
Authors and Affiliations
Contributions
A.S.W.D., F.H., J.M.L., and M.P. conceived of the study. A.S.W.D., L.P., S.M.H., G.M.L., A.C.K., M.F.F., and F.H. initiated the study design and helped with implementation. A.S.W.D. and S.C.V. analyzed flow cytometry data, V.V. and R.G. provided statistical expertise and RNAseq data analysis. S.C.V., J.M.L., and M.P. wrote the initial draft of the manuscript, all authors contributed to the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Woodward Davis, A.S., Vick, S.C., Pattacini, L. et al. The human memory T cell compartment changes across tissues of the female reproductive tract. Mucosal Immunol 14, 862–872 (2021). https://doi.org/10.1038/s41385-021-00406-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-021-00406-6
This article is cited by
-
Tissue-resident memory T cells in the urogenital tract
Nature Reviews Nephrology (2022)
-
Mucosal viral infection induces a regulatory T cell activation phenotype distinct from tissue residency in mouse and human tissues
Mucosal Immunology (2022)
-
Tissue-resident immunity in the female and male reproductive tract
Seminars in Immunopathology (2022)