Abstract
Although they globally cause viral gastroenteritis in children, astroviruses are understudied due to the lack of well-defined animal models. While murine astroviruses (muAstVs) chronically infect immunodeficient mice, a culture system and understanding of their pathogenesis is lacking. Here, we describe a platform to cultivate muAstV using air–liquid interface (ALI) cultures derived from mouse enteroids, which support apical infection and release. Chronic muAstV infection occurs predominantly in the small intestine and correlates with higher interferon-lambda (IFN-λ) expression. MuAstV stimulates IFN-λ production in ALI, recapitulating our in vivo findings. We demonstrate that goblet cells and enterocytes are targets for chronic muAstV infection in vivo, and that infection is enhanced by parasite co-infection or type 2 cytokine signaling. Depletion of goblet cells from ALI limits muAstV infection in vitro. During chronic infection, muAstV stimulates IFN-λ production in infected cells and induces ISGs throughout the intestinal epithelium in an IFN-λ-receptor-dependent manner. Collectively, our study provides insights into the cellular tropism and innate immune responses to muAstV and establishes an enteroid-based culture system to propagate muAstV in vitro.
Similar content being viewed by others
Introduction
Human astroviruses (HAstVs) are positive-sense, single-stranded RNA viruses, and major causes of pediatric gastroenteritis worldwide.1,2,3 In healthy individuals, HAstV infections may be asymptomatic or result in self-limited illness. In immunocompromised individuals, systemic spread can result in encephalitis, meningitis, and lethality.4,5,6 Despite their clinical importance, HAstVs have been understudied due to the lack of a well-defined small animal model. Murine astroviruses (muAstVs) are endemic to many mouse facilities, causing acute asymptomatic infections in immunocompetent mice and chronic infection in immunodeficient models.7,8 We reported that muAstV can complement primary immunodeficiency to protect against other enteric viral infections via stimulation of interferon-lambda (IFN-λ) signaling in the epithelium.9 However, the cellular tropism for muAstV and source of IFN-λ to stimulate this antiviral environment remains unclear.
Here, we report that muAstV can both propagate and drive IFN-λ production in air–liquid interface (ALI) cultures derived from murine intestinal enteroids. We also demonstrate that muAstV chronically infects small intestinal goblet cells in immunodeficient mice, consistent with a recent report identifying goblet cells as a target of muAstV in immunocompetent animals;10 however, we also demonstrate enterocytes as target cells of infection in the small intestine. MuAstV replication in ALI is reduced when goblet cells are depleted. Finally, we demonstrate that muAstV infection drives IFN-λ production in goblet cells and enterocytes, resulting in IFN-λ receptor-dependent protective interferon-stimulated gene (ISG) expression in neighboring cells in the intestinal epithelium.
Results
Murine astrovirus preferentially replicates in the small intestine of immunodeficient and immunocompetent mice, independent of the microbiota
Previously, we showed Rag2−/−Il2rg−/− mice bred at Washington University are persistently infected with muAstV, including the novel strain STL5, which drives a state of chronic IFN-λ-mediated intestinal inflammation.9 To define the preferred site(s) of muAstV replication, we quantified total muAstV and STL5 levels along the GI tract and in extraintestinal tissues from naive Rag2−/−Il2rg−/− mice (Fig. S1A). The small intestine showed significantly higher viral loads for total muAstV and STL5 compared to the esophagus and stomach, with intermediate viral levels detected in the proximal colon (Fig. 1A, B). In the small intestine, higher viral loads correlated with increased IFN-λ (Ifnl2/3) expression (Fig. 1C). Consistent with a previous report, we observed modest levels of muAstV in extraintestinal tissues, ~10,000-fold lower than those detected in small intestinal tissues (Fig. S1B), but did not detect STL5 genomes or Ifnl2/3 expression at these sites7 (Fig. S1C, D). MuAstV, STL5, and Ifnl2/3 were undetectable in astrovirus-free Rag2−/−Il2rg−/− mice from Taconic at all sites tested, consistent with our prior report that muAstV potentiates intestinal IFN-λ expression in immunocompromised mice9 (Fig. S1E–G). Treatment of germ-free (GF) or conventionally-housed wild-type (WT) mice with filtered fecal transplant (FFT) material from Rag2−/−Il2rg−/− mice demonstrated similar preferential infection of the small intestine by muAstV, correlated with enhanced IFN-λ expression at this site, supporting a microbiota-independent tissue tropism for muAstV (Fig. 1D–I). Based on these findings, and our prior observation that specific muAstV strains are preferentially associated with the intestinal epithelial fraction of the intestine,9 we hypothesized that muAstV could be cultivated in intestinal enteroids for further characterization.
A, B MuAstV and STL5 genomes quantified in tissues of Rag2−/−Il2rg−/− mice (n = 4–8). C Ifnl2/3 mRNA levels in tissues of Rag2−/−Il2rg−/− mice (n = 4–8). D, E muAstV and STL5 genomes quantified in tissues of germ-free (GF) WT mice 5 days postinoculation with Rag2−/−Il2rg−/− filtered fecal transplant (FFT) material (n = 4–8). F Ifnl2/3 mRNA levels in tissues of GF WT mice 5 days postinoculation with Rag2−/−Il2rg−/− FFT (n = 4–8). G, H MuAstV and STL5 genomes quantified in tissues of conventionally-housed WT mice 5 days postinoculation with Rag2−/−Il2rg−/− filtered fecal transplant (FFT) material (n = 5). I Ifnl2/3 mRNA levels in tissues of WT mice 5 days postinoculation with Rag2−/−Il2rg−/− FFT (n = 5). Results were analyzed using Kruskal–Wallis ANOVA with Dunn’s post test, combined from two independent experiments. ***P < 0.0001; **P < 0.001. Bars indicate mean of all data points.
Human astrovirus, but not murine astrovirus, can be cultivated in 3D enteroids
Human intestinal enteroids (HIEs) have been used to cultivate a variety of enteric viruses including human rotavirus, human norovirus, human enterovirus, and, of greatest relevance, HAstV.11,12,13,14,15 Apical infection of two-dimensional (2D) HIEs by HAstV strain VA1 recently was demonstrated to support robust viral replication and IFN responses.14 In establishing our cultivation system, we first explored the possibility of HAstV infection in three-dimensional (3D) enteroids. We inoculated differentiated 3D enteroids derived from adult human biopsies with HAstV1 at a multiplicity of infection (MOI) of 1. HIEs inoculated with HAstV1 showed a ~20–30-fold increase in viral genomes at 24 h post infection (hpi) compared to HIEs harvested at 1 hpi or which had been inoculated with UV-inactivated virus. They also exhibited increased expression of type I and III IFNs and multiple ISGs (Fig. 2A–F). However, 3D enteroids generated from mouse small intestine inoculated with FFT containing muAstV from Rag2−/−Il2rg−/− mice failed to result in infection or induce IFN responses (Fig. 2G–L). These results indicate that while HAstVs can infect basolaterally, apical access might be crucial for productive muAstV infection.
A–F Relative quantification of HAstV1 genomes, IFNB1, IFNL2/3, OAS2, MX1, or IFI44 mRNA levels in HIEs infected with HAstV1 at an MOI of 1.0 or UV-treated HAstV1 at 1 hpi (input) and 24 hpi (n = 6). G–L Relative quantification of muAstV genomes, Ifnb1, Ifnl2/3, Ifit1, Oas1a, or Stat1 in mouse enteroids inoculated with Rag2−/−Il2rg−/− FFT at 1 hpi (input) and 24 hpi (n = 6). Results were analysed using Kruskal–Wallis ANOVA with Dunn’s post test (A–F) or Mann–Whitney test (G–L), combined from three independent experiments. ***P < 0.0001; **P < 0.001; *P < 0.01. ns = not significant. Bars indicate mean of all data points.
Air–liquid interface (ALI) cultures support murine astrovirus replication
As the 3D enteroids did not support muAstV replication, we utilized the 3D murine enteroid cells to form 2D monolayers on transwells under ALI conditions (Fig. 3A). We added FFT from Rag2−/−Il2rg−/− mice to the top chamber of the transwells to permit apical infection and collected cells at 1 hpi or 24 hpi to quantify muAstV genomes. We observed a >10-fold increase in viral genome copies at 24 hpi (Fig. 3B, C). Moreover, WT mice inoculated with apical, but not basal, media from Rag2−/−Il2rg−/− FFT-infected ALI transwell cultures exhibited muAstV infection at 3 dpi (Fig. 3D–F). These data suggest exclusively apical viral entry and release for muAstV. Chronic muAstV infection in Rag2−/−Il2rg−/− mice is associated with elevated intestinal IFN-λ expression without associated type I IFN induction9 (Fig. 1C). We observed robust induction of multiple ISGs after muAstV infection in ALI cultures (Fig. 3G–I). ALI cultures infected with muAstV recapitulated the in vivo phenomenon of preferential induction of IFN-λ rather than type I IFNs (Fig. 3J, K). While 2D cultures developed from small intestinal enteroids also supported muAstV replication and IFN-λ induction, the levels were lower than those observed in our ALI cultures (Fig. S2A, B). Together, these findings confirm an epithelial cell tropism for muAstV, and demonstrate the utility of ALI cultures as a model to propagate muAstVs for study of physiologically-relevant innate immune induction mechanisms.
A Schematic for ALI culture workflow. B, C Relative quantification of muAstV and STL5 genomes in ALI cultures inoculated with Rag2−/−Il2rg−/− FFT at 1 hpi (input) and 24 hpi (n = 6). D Schematic for administration of ALI-derived muAstV for in vivo infection. E, F muAstV and STL5 genome copies detected in fecal pellets of WT mice 3 dpi with PBS (mock) or media from the bottom chamber (basal media) or top chamber (apical media) of the ALI culture (n = 8). G–K Relative quantification of Ifit1, Oas1a, Stat1, Ifnb1, or Ifnl2/3 in ALI cultures inoculated with Rag2−/−Il2rg−/− FFT at 1 hpi (input) and 24 hpi (n = 6). Results were analyzed using Mann–Whitney test (B, C; G–K) or Kruskal–Wallis ANOVA with Dunn’s post test (E, F), combined from two independent experiments. ***P < 0.0001; **P < 0.001; *P < 0.01. ns = not significant. Bars indicate mean of all data points.
Murine astrovirus infects goblets cells and enterocytes
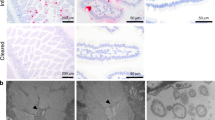
We next explored the specific epithelial cell tropism of muAstV in vivo. First, we performed RNA in situ hybridization on duodenums from naive WT and chronically muAstV-infected Rag2−/−Il2rg−/− mice using muAstV-specific probes. As expected, muAstV RNA was detected only in the Rag2−/−Il2rg−/− mice, and we observed staining exclusively in the villi, supporting an epithelial cell tropism (Fig. S3A). We next costained the tissues with RNA-targeting probes for muAstV and individual epithelial cell surface markers including lysozyme (Lyz-1) for Paneth cells, chromogranin A (ChgA) for enteroendocrine cells, doublecortin-like kinase 1 (Dclk-1) for tuft cells, and mucin 2 (Muc2) for goblet cells. Consistent with our observation of muAstV staining villi, we did not observe colocalization of muAstV-positive cells with Lyz-1, nor did we observe costaining with ChgA or Dclk-1 (Fig. S3B–D). Instead, we observed muAstV infection predominantly in the ACE-2-positive mature enterocytes (Figs. 4A, S3E). Beyond a tropism for mature enterocytes, in Rag2−/−Il2rg−/− mice, ~22% of muAstV-positive cells were Muc2-positive (Figs. 4A, S3E) and ~5% of total goblet cells were targeted for muAstV infection. UEA-1- and cytokeratin-18 (CK-18)-positive goblet cells sorted from Rag2−/−Il2rg−/− small intestines were enriched in muAstV and STL5 genomes, further supporting a goblet cell tropism for muAstV (Figs. 4B, S3F). We also costained intestinal sections with a combination of anti-UEA-1 antibody and the RNA-targeting probe to muAstV, and detected double-positive cells (Fig. S3G). To define the cellular tropism of muAstV in WT mice, we performed RNA in situ hybridization on duodenum sections from WT conventionally-housed and GF mice after FFT. In comparison to Rag2−/−Il2rg−/− mice (3.4 ± 1.7 cells/villus), WT mice had substantially fewer muAstV-positive cells (0.3 ± 0.4 cells/villus). Despite the relative scarcity of these cells, we could detect consistent muAstV and Muc2 costaining, suggesting that a small number of intestinal goblet cell sustain muAstV infection in immunocompetent mice and serve as the predominant site of infection (Figs. 4C, S3H). We did not find robust infection of colonic goblet cells, nor of the colon in general (Fig. S3I), as seen in our qPCR analysis (Fig. 1A). Collectively, our results demonstrate a tropism for goblet cells and enterocytes for muAstV.
A Duodenal sections from WT and Rag2−/−Il2rg−/− mice were hybridized with probes for muAstV (green), Muc2 (red), and anti-ACE-2 antibody (gray). Cell nuclei were stained using DAPI (blue). B Relative quantification of muAstV (left) and STL5 (right) genomes in sorted GCs from small intestines of Rag2−/−Il2rg−/− mice (n = 9). C Duodenal sections from WT and WT GF mice were hybridized with probes for muAstV (green), Muc2 (red), and anti-ACE-2 antibody (gray). Cell nuclei were stained using DAPI (blue). D MuAstV genomes quantified in fecal pellets and tissues of Rag2−/−Il2rg−/− and Rag2−/−Il2rg−/−Ifnlr1−/− mice (n = 5–6). E Duodenal sections from Rag2−/−Il2rg−/−Ifnlr1−/− mice were hybridized with probes for muAstV (green) and Muc2 (red). Cell nuclei were stained using DAPI (blue). Results were analysed using Mann–Whitney test (B, D). ***P < 0.0001; **P < 0.001. Scale bar = 25 µm. Bars indicate mean of all data points. GC Goblet cells.
We previously generated Rag2−/−Il2rg−/−Ifnlr1−/− mice to define the role of IFN-λ in muAstV-driven resistance of immunocompromised mice to other enteric viruses. Experiments with these mice showed that signaling through the receptor for IFN-λ (IL28Rα/Il10Rβ) was critical for this resistance.9 We hypothesized that IFN-λ signaling also might control muAstV levels in chronically infected Rag2−/−Il2rg−/− mice. Indeed, we observed significantly higher muAstV levels in stool and intestinal tissues from Rag2−/−Il2rg−/−Ifnlr1−/− mice compared to Rag2−/−Il2rg−/− (Fig. 4D). Duodenal sections costained with probes for muAstV and Muc2 showed higher numbers of muAstV-positive cells in Rag2−/−Il2rg−/−Ifnlr1−/− mice (11.8 ± 3.9 cells/villus) than Rag2−/−Il2rg−/− mice, with ~12% of total goblet cells infected, suggesting a role for IFN-λ in regulating the number of muAstV-infected cells (Figs. 4E, S3J). The proportion of Muc2-positive infected cells was similar (~23%) in Rag2−/−Il2rg−/−Ifnlr1−/− as in Rag2−/−Il2rg−/− mice. We assessed the total number of Muc2-positive cells, and found they were similar in WT, Rag2−/−Il2rg−/−, and Rag2−/−Il2rg−/−Ifnlr1−/− mice (12.7 ± 5.3, 12.6 ± 2.3, and 16.5 ± 11.4 cells/villus, respectively).
Regulation of goblet cells modulates muAstV infection
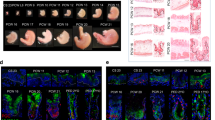
In the intestinal epithelium, the abundance of secretory cells including goblet cells is tightly regulated. Gastrointestinal nematodes have been shown to induce severe cellular hyperplasia characterized by rapid expansion of goblet, tuft and Paneth cells upon infection.16,17,18 Infection of WT mice with murine nematode parasite Heligmosomoides polygyrus (H. polygyrus) prior to Rag2−/−Il2rg−/− FFT significantly enhanced muAstV infection (Figs. 5A, S4A). Similarly, treatment with IL-4 or IL-33, known drivers of goblet cell hyperplasia19,20 that stimulate increased expression of Muc2 in the intestine, resulted in a marked increase in muAstV infection in mice administered Rag2−/−Il2rg−/− FFT (Figs. 5B, C, S4B–D), suggesting that increased goblet cell numbers enhance muAstV infection. We did not detect higher expression of Muc2 with Rag2−/−Il2rg−/− FFT treatment of WT mice (Fig. S4D). To test the requirement for goblet cells in muAstV infection, we generated small intestinal enteroids from Math1f/f-Vil-Cre-ERT2 mice, which were previously used for inducible tamoxifen-mediated depletion of goblet cells in vivo.21 To adapt this model in vitro, we confirmed the capacity of enteroids from Math1f/f-Vil-Cre-ERT2 to differentiate into secretory cell lineages in the absence of tamoxifen (Fig. S4E). Upon treatment with 4-hydroxytamoxifen (4-OHT), we observed a marked reduction in the expression of secretory cell markers Muc2, ChgA and Lyz-1, although the stem cell marker Lgr5 remained unchanged (Fig. 5D). Depletion of goblet cells at 72 h after 4-OHT treatment was confirmed by staining of enteroids with anti-UEA-1 and anti-ChgA (Fig. 5E), and 4-OHT treatment of ALI cultures from Math1f/f-Vil-Cre-ERT2 mice similarly reduced Muc2 levels (Figs. 5F, S4F). MuAstV infection at 48 hpi was significantly inhibited in 4-OHT-treated cultures compared to vehicle-treated controls (Fig. 5F). These results suggest that goblet cells are necessary for maximal replication of muAstV, and that goblet cell abundance regulates muAstV infection in vivo and in vitro. Furthermore, induction of Ifnl2/3 was significantly reduced in 4-OHT-treated ALI (Fig. 5F), suggesting goblet cells might be a source of IFN-λ during muAstV infection.
A MuAstV genome copies in stool of WT mice 5 days postinoculation with Rag2−/−Il2rg−/− FFT, following prior infection with H. polygyrus or mock (n = 8–10). B, C MuAstV genome copies detected in stool and ileum of WT mice administered PBS, IL-4c, or IL-33 prior to FFT treatment (n = 5–6). D Relative quantification of mRNA levels of intestinal cell markers Lgr5, Muc2, ChgA, and Lyz-1 in Math1f/f-Vil-Cre-ERT2 organoids treated with ethanol (vehicle) or 300 nM 4-OHT for 48 h. E Math1f/f-Vil-Cre-ERT2 organoids treated with ethanol or 4-OHT stained for anti-UEA-1 (green) and anti-ChgA (red). Cell nuclei were stained using DAPI (blue). F Relative quantification of Muc2, muAstV, and Ifnl2/3 levels at 2dpi in Math1f/f-Vil-Cre-ERT2-derived ALI cultures treated with ethanol (vehicle) or 4-OHT for 3 days prior to Rag2−/−Il2rg−/− FFT inoculation. Results analysed using Mann–Whitney test (A, D) and Kruskal–Wallis ANOVA with Dunn’s post test (B, C, F), from two independent experiments. ***P < 0.0001; **P < 0.001. Scale bar = 25 µm. Bars indicate mean of all data points.
MuAstV infection stimulates IFN-λ expression in small intestinal goblet cells and enterocytes to drive widespread ISG expression
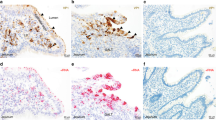
To examine further the source of IFN-λ expression in the intestine, we performed RNA in situ hybridization on intestinal sections of naive WT, WT treated with Rag2−/−Il2rg−/− FFT and Rag2−/−Il2rg−/− mice using probes for Ifnl2/3. This analysis revealed increased Ifnl2/3 staining in WT mice treated with Rag2−/−Il2rg−/− FFT (0.2 ± 0.1 cells/villus) and Rag2−/−Il2rg−/− mice (0.7 ± 0.2 cells/villus) compared to naive WT mice (0 cells/villus) (Figs. 6A, S5A). In both WT and Rag2−/−Il2rg−/− mice treated with FFT, Ifnl2/3-positive cells colocalized with muAstV-positive cells, indicating that muAstV-infected cells directly produce IFN-λ (Figs. 6A, S5A). The Ifnl2/3-positive cells observed in the Rag2−/−Il2rg−/− mice were predominantly Muc2-negative, suggesting that infected enterocytes preferentially secrete IFN-λ in these animals (Figs. 6A, S5B). Rag2−/−Il2rg−/−Ifnlr1−/− mice exhibited greater intestinal Ifnl2/3 expression compared to Rag2−/−Il2rg−/− mice, perhaps secondary to elevated levels of muAstV and/or a lack of negative feedback signaling in the absence of an IFN-λ response (Fig. 6B). Consistent with this expression data, we observed more Ifnl2/3-positive cells in Rag2−/−Il2rg−/−Ifnlr1−/− mice (4.0 ± 0.3 cells/villus) (Figs. 6C, S5B). In the Rag2−/−Il2rg−/−Ifnlr1−/− mice, we observed colocalization of Ifnl2/3-positive cells with both Muc2-positive and Muc2-negative cells, indicating that goblet cells have the capacity for IFN-λ production (Figs. 6C, S5B). Corresponding increases in protein levels of IFN-λ in the serum and small intestine of Rag2−/−Il2rg−/− and Rag2−/−Il2rg−/−Ifnlr1−/− mice were confirmed by ELISA (Fig. 6D). We performed RNA in situ hybridization for the ISG Ifit1 to determine which epithelial cells responded to the elevated levels of IFN-λ in our immunocompromised lines. We observed widespread ISG expression throughout the epithelium, indicating that IFN-λ-stimulated ISGs are expressed both in infected and neighboring cells (Figs. 6E, S5C). This widespread ISG expression was not detected in the naive WT duodenum, and as expected, was blunted in Rag2−/−Il2rg−/−Ifnlr1−/− mice, which lack the capacity to respond to IFN-λ (Figs. 6E, S5C). We confirmed that ISG expression in goblet cells was similar to other epithelial cells by analyzing sorted cells (Fig. S5D). Collectively, our results identify goblet cells and enterocytes as cellular targets for muAstV in immunodeficient mice, and demonstrate infected epithelial cells, including goblet cells and enterocytes, are sources of IFN-λ during chronic infection. We further clarify that this IFN-λ response drives ISG expression throughout the epithelium, thus explaining the resistance of our Rag2−/−Il2rg−/− mice to enteric infections with different epithelial cell targets.9
A Duodenal sections from WT, WT treated with Rag2−/−Il2rg−/− FFT and Rag2−/−Il2rg−/− mice were hybridized with probes for muAstV (green), Ifnl2/3 (red or green), and Muc2 (red). Cell nuclei were stained using DAPI (blue). B Relative quantification of Ifnl2/3 mRNA levels in ileum and colon of Rag2−/−Il2rg−/− and Rag2−/−Il2rg−/−Ifnlr1−/− mice. C Duodenal sections from Rag2−/−Il2rg−/− Ifnlr1−/− mice were hybridized with probes for Muc2 (red) and Ifnl2/3 (green). Cell nuclei were stained using DAPI (blue). D IFN-λ protein levels in serum (left) and small intestine (right) of WT, Rag2−/−Il2rg−/−, and Rag2−/−Il2rg−/−Ifnlr1−/− mice quantified by ELISA. E Duodenal sections from WT, Rag2−/−Il2rg−/−, and Rag2−/−Il2rg−/− Ifnlr1−/− mice were hybridized with probes for muAstV (green) and Ifit1 (red). Cell nuclei were stained using DAPI (blue). Results were analyzed using Mann–Whitney test (B, D), from two independent experiments. **P < 0.001; *P < 0.01. Scale bar = 25 µm.
Discussion
In this study, we investigated the tissue and cellular tropism of muAstV during chronic and acute infection of immunodeficient and immunocompetent mice, respectively. Consistent with a recent report defining goblet cells as a key target for muAstV in immunocompetent mice,10 we identified small intestinal goblet cells as a site for muAstV infection. Along with goblet cells, we also demonstrated that mature enterocytes are a major target for muAstV infection in immunocompromised mice. Goblet cells play a key role in maintaining the intestinal barrier through secretion of mucus, antimicrobial proteins, chemokines, and cytokines to control bacterial and viral pathogens.22 To assess the consequences of modulation of goblet cells on muAstV infection, we infected mice with nematode parasite H. polygyrus which targets the small intestine, a key site of muAstV infection. H. polygyrus infection induces a type 2 immune response, which increases levels of cytokines IL-4, IL-5, IL-9, and IL-13, and stimulates goblet cell hyperplasia in WT mice.23,24 H. polygyrus infection resulted in increased muAstV replication. Recombinant IL-4 and IL-33 treatment also stimulate intestinal goblet cell hyperplasia,20,25 and similarly enhanced muAstV replication in vivo.
Math1 has been reported previously to control the development of secretory cell lineages including Paneth cells, goblet cells and enteroendocrine cells.26 Recently, inducible depletion of goblet cells was demonstrated by specifically deleting Math1 in intestinal epithelial cells.27 We harnessed this model to demonstrate that depletion of goblet cells in vitro reduces muAstV infection. Thus, our findings indicate that either positive or negative modulation of goblet cell numbers correspondingly regulates muAstV infection. Exploration of whether the presence of goblet cells themselves or goblet cell products such as mucus help regulate muAstV infection will be a key future avenue of study.
Our findings in immunocompetent mice are broadly concordant with a recent report of goblet cells being the main target cell type for muAstV.10 However, in immunocompromised mice we observed that while goblet cells were infected, mature enterocytes were the predominant site of infection, suggesting an expanded tropism during chronic infection. Our findings agree with the multicellular tropism reported for human AstV, including mature enterocytes, goblet cells, and intestinal progenitor cells.14 Similarly, while IL-4 treatment was not observed to enhance muAstV infection in a prior study,10 viral strain, timing and facility differences may govern variability in the relative proportions of intestinal epithelial cell types, as well as the sensitivity of these cell types to cytokine-driven modulation, that are infected by muAstV.
We demonstrated that muAstV infection preferentially stimulates IFN-λ production during infection. We also showed infected goblet cells and enterocytes are sources of IFN-λ upon muAstV infection, driving ISG expression throughout the epithelium. Rag2−/−Il2rg−/− mice are resistant to murine norovirus, which has a tropism for tuft cells,28 and murine rotavirus, which has a tropism for mature enterocytes.29 The widespread response to IFN-λ throughout the epithelium explains this broad protection. Robust IFN-λ induction is not a universal feature of muAstV infection, as we previously reported that chronically infected Rag1−/− mice in our facility with a different muAstV strain composition exhibit WT levels of intestinal ISGs.9 Further study of the effects of strain and cellular tropism on IFN-λ induction will be critical to better understand these differences. While we did not find increased Muc2 expression with muAstV infection in vitro or in vivo, muAstV infection has been reported to increase mucus production in the small intestine, mediating protection against enteropathogenic E. coli.10 It is possible that enhanced mucus production could also contribute to protection of muAstV-infected mice against other viruses. Future work exploring the changes in gene expression with muAstV infection in the epithelial cell lineages may provide insights into cell-specific innate immune responses during infection.
We also report the successful propagation of muAstVs in vitro using ALI and 2D cultures but not 3D enteroids. Our studies in this system indicate apical viral entry and egress. Interrogation of the specific host factors that are appropriated by muAstV for these processes, and whether these are associated with any canonical functions of the intestinal epithelium such as mucus production or nutrient absorption, will be important future areas of investigation. Our ALI cultures additionally reflected the in vivo response to muAstV infection with type III, but not type I, IFN expression. Our study highlights the importance of this system for further study of astrovirus infections of the intestinal epithelium and the innate immune responses they elicit.
Materials and methods
Mice
WT C57BL/6J (stock no. 000664) mice were purchased from Jackson Laboratories and maintained at Washington University School of Medicine under specific-pathogen-free conditions according to University guidelines. Rag2−/−Il2rg−/− mice were generated at Washington University School of Medicine by crossing of Il2rg−/− (B6.129S4-Il2rgtm1Wjl/J, catalogue no. 003174) mice from Jackson Laboratories with Rag2−/− (B6.129S6-Rag2tm1Fwa N12, catalogue no. RAGN12) mice from Taconic. Rag2−/−Il2rg−/−Ifnlr1−/− mice were generated as previously described.9 Rag2−/−Il2rg−/− mice from Taconic were used directly for experiments on arrival at Washington University facilities. Math1f/f mice were crossed to transgenic mice bearing a tamoxifen-dependent Cre recombinase expressed under the control of the Villin promoter (Vil-Cre-ERT2)30 to generate Math1f/f-Vil-Cre-ERT2 mice as previously described.21 Germ-free (GF) Swiss Webster mice were purchased from Taconic Biosciences and maintained in sterile, flexible-film plastic gnotobiotic isolators at the Washington University School of Medicine Gnotobiotic Facility. Equal ratios of adult male and female mice, aged 6–12 weeks, were used in all experiments for all strains. Experimental mice were co-housed with up to five mice of the same sex per cage with autoclaved standard chow pellets and water provided ad libitum.
Human and mouse enteroid culture
Human intestinal tissue biopsies were obtained with approval from the Washington University in St. Louis institutional review board (no. 201804040). The HIEs were generated as described previously.31 Briefly, tissue sample was digested in collagenase I (Invitrogen, Waltham, MA) for 10 min at 37 °C followed by mechanical dissociation. The resulting crypts were filtered through a 70 μm cell strainer and washed in Dulbecco’s modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum before resuspending in growth factor-reduced Matrigel (Becton Dickinson, Franklin Lakes, NJ), and seeded into culture plates. Organoids were grown in 50% L-WRN conditioned medium supplemented with 10 μM Y-27632 and SB 202190 (R&D Systems).
Murine enteroids were generated from crypts isolated from small intestines. Briefly, adult mice were euthanized to collect small intestinal tissue. The tissue was cut into 2–4 mm pieces and washed with ice cold PBS. The tissue fragments were treated with EDTA solution to isolate intestinal crypts. The crypts were resuspended in Matrigel and seeded into culture plates. The organoids were grown in basal culture media [Advanced DMEM/F12 (Gibco) supplemented with 2 mM GlutaMax (Invitrogen), 10 mM HEPES]. For differentiation, the organoids were cultured in differentiation media consisting of basal culture medium with (1X) N2 supplement (Invitrogen), (1X) B27 supplement (Invitrogen), and 1 mM N-acetylcysteine (Sigma Aldrich), 50 ng/mL murine recombinant EGF (Invitrogen), 100 ng/mL murine recombinant Noggin (PeproTech), 1 mg/mL human recombinant R-spondin 1 (R&D Systems).32 For depletion of goblet cells, organoids cultured in differentiation media were treated with 300 nM 4-OHT (Sigma Aldrich) for 48 h as described previously.33 Small intestinal enteroids were dissociated into single cells by trypsin treatment and 1 × 105 cells were seeded in L-WRN conditioned media in 48-well plates to form 2D monolayers. The 2D monolayers were maintained in L-WRN media and allowed to form a confluent monolayer. For differentiation, the 2D monolayers were cultured in differentiation media for up to 6 days.
ALI culture and infections
Small intestinal enteroids were dissociated into single cells by 0.25% trypsin digestion prior to seeding into Transwells with 0.4 μm pore size (Corning) precoated with 10% Matrigel (Corning) for 15 min. Cells were cultured in L-WRN conditioned media with 10 μM Rock inhibitor Y-27632 (R&D Systems).34,35 After 7 days of culture, medium from the upper chamber of the transwells was removed to create an ALI. Cells were maintained in ALI in L-WRN conditioned media with 10 μM Rock inhibitor Y-27632 in the bottom chamber for 14 days while changing the media every 2 days. For goblet cell depletion, ALI cultures were propagated in 300 nM of 4-OHT supplemented in L-WRN conditioned media with 10uM Rock inhibitor Y-27632 for 72 h prior to infection.
For infection of ALI and 2D enteroid cultures, differentiated cultures were treated with FFT from Rag2−/−Il2rg−/− mice diluted in PBS for 1 h. The inoculum was removed and culture wells were washed with PBS twice before collecting the 1 hpi timepoint in Tri Reagent. For 2D cultures, fresh differentiation media was added, and the cells were allowed to grow for 48 h. For ALI cultures, fresh 50% L-WRN conditioned media was added to the top and bottom chambers and cells were allowed to grow for 24 or 48 h. For propagating muAstV, FFT from Rag2−/−Il2rg−/− mice was added to the top chamber of ALI culture transwells. After 1 h, both top and bottom chamber were washed with PBS twice. Fresh L-WRN conditioned media was added to the top and bottom chamber and cells were allowed to grow for 2 days. After 2 days, media from the top and bottom chamber was collected to orally gavage WT adult mice.
Virus infections
For muAstV infection, mice were administered FFT as described previously.9 For muAstV infection in enteroids, differentiated mouse 3D enteroids were incubated with FFT diluted in PBS for 1 h followed by washing with PBS twice. For 1 hpi timepoint, the enteroids were directly harvested in Tri Reagent (Invitrogen). For viral replication, the enteroids were cultured in L-WRN conditioned media supplemented with 10 μM Y-27632 and 10 μM SB431542.
Human astrovirus strain HAstV1 was grown in Caco2 cells as described previously.36 For HAstV1 infection in human 3D enteroids, differentiated enteroids were incubated with HAstV1 in PBS for 1 h. The enteroids were washed with PBS twice and 1 hpi samples were harvested in Tri reagent. The HAstV1-infected enteroids were grown in L-WRN conditioned media supplemented with 10 μM Y-27632 and 10 μM SB431542.
H. polygyrus infection
Heligmosomoides polygyrus bakeri (H. polygyrus) third-stage larvae (L3) were generated as described.37 H.polygyrus L3 viability was checked by microscope for motility, and their numbers were quantified before use. Mice were gavaged with 200 H.polygyrus L3 or PBS (control) using 20-gauge × 38 mm plastic feeding tubes (Instech; FTP-20-38) prior to infection with muAstV.
Cytokine administration
Recombinant murine IL-4 complexes (IL-4c) were generated as described previously.38 Briefly, 5 μg of murine IL-4 (Peprotech) was mixed with 25 μg of anti-IL-4 (BioXCell) and incubated for 15 min prior to diluting in PBS to a final volume of 200 μl. This mixture of IL-4c was administered via intraperitoneal injection to each mouse at 48-h intervals on day −2 and 0 prior to treatment with FFT. Recombinant IL-33 (Peprotech) was diluted to 0.4 μg per mouse in 200 μl of PBS and administered via intraperitoneal injection on day −2 and 0 before treatment with FFT, and day 2 after treatment with FFT as shown in Fig. S3C.
RNA extraction, quantitative reverse transcription-PCR, and enzyme-linked immunosorbent assay (ELISA)
Total RNA from stool was isolated using a ZR-96 Viral RNA kit (Zymo Research). Total RNA from tissues or cells was isolated using Tri Reagent (Invitrogen) and a Direct-zol-96 RNA kit (Zymo Research) according to the manufacturer’s protocol. ImPromII reverse transcriptase was used for cDNA synthesis (Promega). qPCR for muAstV, designed to detect all muAstV strains using primers against conserved regions, was performed as described previously.7 A SYBR green qPCR assay was performed for muAstV STL5 as described previously.9 HAstV1 was quantified using primers specific for HAstVs [Mon269 (5′-CAACTCAGGAAACAGGGTGT-3′) and Mon270 (5′-TCAGATGCATTGTCATTGGT-3′)].39 Predesigned PrimeTime qPCR assays were used to quantify expression of human and mouse genes, with assays used in this study listed in Table 1. SYBR green qPCR assays were performed with Power SYBR Green PCR master mix (ThermoFisher) using primers listed in Table 2.
For IFN-λ ELISA, serum or 30–50 mg of small intestinal homogenate were collected and IFN-λ2/IFN-λ3 levels were analyzed by ELISA as described previously (R&D Systems).40
RNAscope in situ hybridization
For preparing tissue sections, duodenum Swiss rolls were fixed overnight in 10% formalin then immersed in freshly prepared 30% sucrose solution until the tissue sunk to the bottom of the tube (48 h). Tissues were then frozen in Optimal Cutting Temperature (OCT) embedding media over dry ice. Fixed frozen tissue sections of 15 µm thickness were then dried at −20 °C for 2 h or kept at −80 °C if not processed immediately. Slides were washed by dipping in 1X PBS for 5 min, followed by a baking step at 60 °C for 30 min, then sections were post fixed by immersing in prechilled 10% formalin for 15 min at 4 °C followed by a dehydration step by immersing slides in a series of increasing ethanol concentrations (50, 70, and 100%) for 5 min each. All RNAscope ISH assays were performed using the RNAscope 2.5 HD Assay-BROWN kit according to manufacturer’s instructions (Advanced Cell Diagnostics, Newark, CA). In brief, tissues were hybridized with custom-designed probes specific for murine astrovirus STL1 RNA for 2 h at 40 °C. A positive control probe specific to the PPIB housekeeping gene transcript and a negative control probe specific to the bacterial DapB transcript were included in each experiment. Sections were then counterstained with 50% hematoxylin to visualize cellular architecture. Images were acquired using an AxioScan Z1 (Zeiss) slide scanner and ZEN Digital Imaging software (Zeiss) was used to analyze the images. Images are representative of two independent experiments.
RNAscope fluorescence in situ hybridization and histology
The RNAscope Multiplex Fluorescent Detection Kit v2 (Advanced Cell Diagnostics, Newark, CA) was used to perform dual hybridizations with murine astrovirus probes in combination with either Ifnl2/3, Ifit1, or cell-specific probes (Muc2 for goblet cells, ChgA for enteroendocrine cells, Dclk-1 for tuft cells, and Lyz-1 for Paneth cells).41 Fixed frozen sections were prepared as in ISH, and signal was detected using fluorescent Opal dyes (Akoya Biosciences). Opal 520 was used for astrovirus probes and Opal 570 was used for Ifnl2/3 as well as the cell-specific markers. For histological analysis, the tissues were stained with primary antibodies overnight at 4 °C, and detected by Alexa Fluor 594 or 647-conjugated secondary antibodies (Invitrogen). Tissues were counterstained with DAPI and mounted using VectaMount mounting medium (Vector Laboratories). Images for FISH were acquired using an ApoTome microscope (Zeiss) and images were captured using Axiovision 4.8.2 software (Zeiss). Single color controls, along with positive and negative control probes (PPIB and DapB, respectively) were stained in parallel. Images are representative of two independent experiments. Primary antibodies used: UEA-1-FITC (Sigma Aldrich), ACE-2 (R&D Systems).
Flow cytometric sorting of goblet cells
The epithelial fraction of the small intestine was isolated as described previously.9 Briefly, mice were euthanized, and small intestine was collected in cold PBS. Tissues were washed with cold PBS twice, chopped and transferred to new tubes followed by incubation in stripping buffer (10% FBS, 15 mM HEPES, 5 mM EDTA, in 1X HBSS) for 20 min at 37 °C. The tissues were vortexed for 15 s and passed sequentially through 100 and 40 μm filters. The collected cells were washed with cold PBS and stained with live/dead staining kit (live/dead fixable blue dead cell stain kit, Life Technologies) according to manufacturer’s protocol. After live/dead staining, cells were stained with anti-CD24 (eBioscience; clone 30-F1), anti-CD45 (Biolegend, clone), anti-UEA-1 (Vector labs), anti-cytokeratin-18 (Abcam; clone C-04). Goblet cells were sorted using a BD ARIA II cell sorter (BD Biosciences) by gating on CD45−CD24−CK-18+UEA-1+, while the remaining IECs were collected by gating on CD45−CD24−CK-18−UEA-1– and collected in RPMI containing 10% FBS.21 The collected cells were centrifuged and resuspended in Tri Reagent (Invitrogen) for RNA extraction. Data were processed using FACSDiva software (BD) and FlowJo (FlowJo).
Data availability
The data from this study are tabulated in the main paper and Supplementary Materials. All reagents are available from M.T.B. under a material transfer agreement with Washington University.
References
Wohlgemuth, N., Honce, R. & Schultz-Cherry, S. Astrovirus evolution and emergence. Infect. Genet. Evol. 69, 30–37, https://doi.org/10.1016/j.meegid.2019.01.009 (2019).
Bosch, A., Pinto, R. M. & Guix, S. Human astroviruses. Clin. Microbiol. Rev. 27, 1048–1074, https://doi.org/10.1128/CMR.00013-14 (2014).
Collaborators, G. B. D. D. D. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 18, 1211–1228, https://doi.org/10.1016/S1473-3099(18)30362-1 (2018).
Wunderli, W. et al. Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PLoS ONE 6, e27483, https://doi.org/10.1371/journal.pone.0027483 (2011).
Quan, P. L. et al. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg. Infect. Dis. 16, 918–925, https://doi.org/10.3201/eid1606.091536 (2010).
Lum, S. H. et al. An emerging opportunistic infection: fatal astrovirus (VA1/HMO-C) encephalitis in a pediatric stem cell transplant recipient. Transpl. Infect. Dis. 18, 960–964, https://doi.org/10.1111/tid.12607 (2016).
Yokoyama, C. C. et al. Adaptive immunity restricts replication of novel murine astroviruses. J. Virol. 86, 12262–12270, https://doi.org/10.1128/JVI.02018-12 (2012).
Cortez, V. et al. Characterizing a murine model for astrovirus using viral isolates from persistently infected immunocompromised mice. J. Virol. 93, https://doi.org/10.1128/JVI.00223-19 (2019).
Ingle, H. et al. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-lambda. Nat. Microbiol. 4, 1120–1128, https://doi.org/10.1038/s41564-019-0416-7 (2019).
Cortez, V. et al. Astrovirus infects actively secreting goblet cells and alters the gut mucus barrier. Nat. Commun. 11, 2097, https://doi.org/10.1038/s41467-020-15999-y (2020).
Drummond, C. G. et al. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc. Natl Acad. Sci. U.S.A. 114, 1672–1677, https://doi.org/10.1073/pnas.1617363114 (2017).
Ettayebi, K. et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393, https://doi.org/10.1126/science.aaf5211 (2016).
Saxena, K. et al. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J. Virol. 90, 43–56, https://doi.org/10.1128/JVI.01930-15 (2016).
Kolawole, A. O. et al. Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape. PLoS Pathog. 15, e1008057, https://doi.org/10.1371/journal.ppat.1008057 (2019).
Good, C., Wells, A. I. & Coyne, C. B. Type III interferon signaling restricts enterovirus 71 infection of goblet cells. Sci. Adv. 5, eaau4255, https://doi.org/10.1126/sciadv.aau4255 (2019).
Coakley, G. & Harris, N. L. The intestinal epithelium at the forefront of host-helminth interactions. Trends Parasitol. 36, 761–772, https://doi.org/10.1016/j.pt.2020.07.002 (2020).
Belle, N. M. et al. TFF3 interacts with LINGO2 to regulate EGFR activation for protection against colitis and gastrointestinal helminths. Nat. Commun. 10, 4408, https://doi.org/10.1038/s41467-019-12315-1 (2019).
Madden, K. B. et al. Enteric nematodes induce stereotypic STAT6-dependent alterations in intestinal epithelial cell function. J. Immunol. 172, 5616–5621, https://doi.org/10.4049/jimmunol.172.9.5616 (2004).
Horsnell, W. G. et al. Delayed goblet cell hyperplasia, acetylcholine receptor expression, and worm expulsion in SMC-specific IL-4Ralpha-deficient mice. PLoS Pathog. 3, e1, https://doi.org/10.1371/journal.ppat.0030001 (2007).
Mahapatro, M. et al. Programming of intestinal epithelial differentiation by IL-33 derived from pericryptal fibroblasts in response to systemic infection. Cell Rep. 15, 1743–1756, https://doi.org/10.1016/j.celrep.2016.04.049 (2016).
Knoop, K. A., McDonald, K. G., McCrate, S., McDole, J. R. & Newberry, R. D. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol. 8, 198–210, https://doi.org/10.1038/mi.2014.58 (2015).
Knoop, K. A. & Newberry, R. D. Goblet cells: multifaceted players in immunity at mucosal surfaces. Mucosal Immunol. 11, 1551–1557, https://doi.org/10.1038/s41385-018-0039-y (2018).
Finkelman, F. D. et al. Cytokine regulation of host defense against parasitic gastrointestinal nematodes: lessons from studies with rodent models. Annu. Rev. Immunol. 15, 505–533, https://doi.org/10.1146/annurev.immunol.15.1.505 (1997).
Hashimoto, K. et al. Depleted intestinal goblet cells and severe pathological changes in SCID mice infected with Heligmosomoides polygyrus. Parasite Immunol. 31, 457–465, https://doi.org/10.1111/j.1365-3024.2009.01123.x (2009).
Gerbe, F. et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230, https://doi.org/10.1038/nature16527 (2016).
Shroyer, N. F. et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 132, 2478–2488, https://doi.org/10.1053/j.gastro.2007.03.047 (2007).
Kulkarni, D. H. et al. Goblet cell associated antigen passages are inhibited during Salmonella typhimurium infection to prevent pathogen dissemination and limit responses to dietary antigens. Mucosal Immunol. 11, 1103–1113, https://doi.org/10.1038/s41385-018-0007-6 (2018).
Wilen, C. B. et al. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science 360, 204–208, https://doi.org/10.1126/science.aar3799 (2018).
Greenberg, H. B. & Estes, M. K. Rotaviruses: from pathogenesis to vaccination. Gastroenterology 136, 1939–1951, https://doi.org/10.1053/j.gastro.2009.02.076 (2009).
el Marjou, F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193, https://doi.org/10.1002/gene.20042 (2004).
Sugimoto, S. & Sato, T. Establishment of 3D intestinal organoid cultures from intestinal stem cells. Methods Mol. Biol. 1612, 97–105, https://doi.org/10.1007/978-1-4939-7021-6_7 (2017).
Sato, T. & Clevers, H. Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 945, 319–328, https://doi.org/10.1007/978-1-62703-125-7_19 (2013).
Wiener, Z. et al. Oncogenic mutations in intestinal adenomas regulate Bim-mediated apoptosis induced by TGF-beta. Proc. Natl Acad. Sci. U.S.A. 111, E2229–E2236, https://doi.org/10.1073/pnas.1406444111 (2014).
Miyoshi, H. & Stappenbeck, T. S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8, 2471–2482, https://doi.org/10.1038/nprot.2013.153 (2013).
Wang, Y. et al. Long-term culture captures injury-repair cycles of colonic stem cells. Cell 179, 1144–1159.e15, https://doi.org/10.1016/j.cell.2019.10.015 (2019).
Marvin, S., Meliopoulos, V. & Schultz-Cherry, S. Human astrovirus propagation, purification and quantification. Bio-Protocol 4, e1078 (2014).
Camberis, M., Le Gros, G. & Urban, J. Jr. Animal model of nippostrongylus brasiliensis and Heligmosomoides polygyrus. Curr. Protoc. Immunol. Chapter 19, 12, https://doi.org/10.1002/0471142735.im1912s55 (2003).
Finkelman, F. D. et al. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J. Immunol. 151, 1235–1244 (1993).
Noel, J. S., Lee, T. W., Kurtz, J. B., Glass, R. I. & Monroe, S. S. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J. Clin. Microbiol. 33, 797–801 (1995).
Peterson, S. T. et al. Disruption of type III interferon genes Ifnl2 and Ifnl3 recapitulates loss of the type III IFN receptor in the mucosal antiviral response. J. Virol. https://doi.org/10.1128/JVI.01073-19 (2019).
Jee, J. H. et al. Development of collagen-based 3D matrix for gastrointestinal tract-derived organoid culture. Stem Cells Int. 2019, 8472712, https://doi.org/10.1155/2019/8472712 (2019).
Acknowledgements
We acknowledge all members of the Baldridge laboratory for helpful discussions. We also thank H. Deng for animal care and breeding, and J. White and the Washington University Gnotobiotic Facility for assistance with germ-free mice. We are grateful to Stacey Schultz-Cherry for providing human astrovirus. H.I. was supported by the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital Postdoctoral Research grant (MI-F-2018-712). S.L. was supported by NIH grant R00 AI141683 and the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (NRF-2016R1A6A3A03012352). M.S.D. was supported by NIH grant R01 DK122790. M.T.B. was supported by NIH grants R01 AI127552, R01 AI139314, the G. Harold and Leila Y. Mathers Foundation, and the Pew Biomedical Scholars Program of The Pew Charitable Trusts.
Author information
Authors and Affiliations
Contributions
H.I., E.H., M.B., Y.L., E.A.K., G.K., H.M., P.D. performed the experiments. H.I., E.H., J.G., M.B., Y.L., H.M., S.L., M.G., M.S.D., and M.T.B. analyzed the results. K.G.M., M.S.D., and R.D.N. provided reagents and helped to design experiments. H.I., M.G., and M.T.B. designed the project. H.I., E.H., J.G., and M.T.B. wrote the paper. All authors read and edited the paper.
Corresponding author
Ethics declarations
Competing interests
M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, Carnival Corporation, and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
About this article
Cite this article
Ingle, H., Hassan, E., Gawron, J. et al. Murine astrovirus tropism for goblet cells and enterocytes facilitates an IFN-λ response in vivo and in enteroid cultures. Mucosal Immunol 14, 751–761 (2021). https://doi.org/10.1038/s41385-021-00387-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-021-00387-6
This article is cited by
-
The impact of helminth-induced immunity on infection with bacteria or viruses
Veterinary Research (2023)
-
The role of goblet cells and mucus in intestinal homeostasis
Nature Reviews Gastroenterology & Hepatology (2022)