Abstract
Induction of intestinal T helper cell subsets by commensal members of the intestinal microbiota is an important component of the many immune adaptations required to establish host-microbial homeostasis. Importantly, altered intestinal T helper cell profiles can have pathological consequences that are not limited to intestinal sites. Therefore, microbial-mediated modulation of the intestinal T helper cell profile could have strong therapeutic potentials. However, in order to develop microbial therapies that specifically induce the desired alterations in the intestinal T helper cell compartment one has to first gain a detailed understanding of how microbial composition and the metabolites derived or induced by the microbiota impact on intestinal T helper cell responses. Here we summarize the milestone findings in the field of microbiota-intestinal T helper cell crosstalk with a focus on the role of specific commensal bacteria and their metabolites. We discuss mechanistic mouse studies and are linking these to human studies where possible. Moreover, we highlight recent advances in the field of microbial CD4 T cell epitope mimicry in autoimmune diseases and the role of microbially-induced CD4 T cells in cancer immune checkpoint blockade therapy.
Similar content being viewed by others
Introduction
Intestinal T helper cell adaptions
At birth, our intestinal tract is successively colonized by microbes, primarily commensal bacteria but also fungi, protozoa, parasites, and viruses. The intestinal microbiota has co-evolved with the immune system1 to maintain a symbiotic relationship and aids in the digestion of inaccessible nutrients, provides colonization resistance to pathogens, and educates the immune system.2 A dynamic crosstalk between the microbiota and the host immune system during compositional or metabolic changes in the microbiota is essential for maintaining local and systemic immune homeostasis. The induction of distinct CD4 T cell subsets in mucosal tissues represents a key adaption in response to bacterial colonization. The intestinal CD4 T cell compartment is composed of functionally diverse subsets with T helper type 1 (Th1), Th2, Th17, T follicular helper (TFH), and regulatory T (Treg) cells being the most prominent and best characterized populations in the intestine. The modulation of T cell function by intestinal commensal bacteria and their metabolites and its consequences on health and disease is the main topic of this review. Other commensal microbes such as fungi have also been shown to modulate intestinal immunity during homeostasis or inflammation.3,4,5,6,7 However, our review will focus on the effects mediated by commensal bacteria.
Bacterial metabolites
Immunomodulatory bacterial metabolites can travel systemically and ultimately influence health and disease at extra-intestinal sites.8,9,10 The importance of bacterial metabolites on host biology was initially highlighted by the comparison of the metabolic profile of germ-free and conventionally housed animals.8,9 These studies demonstrated drastic alterations in the plasma, urine, liver, kidney, and gut tissue metabolomes. More recently, Uchimura et al. utilized stable isotope tracing to demonstrate that microbial metabolites broadly penetrate host systemic organs and induce widespread host metabolic and immunological responses.10
Immune-mediated diseases
Autoimmune and inflammatory diseases that are driven by an imbalance in intestinal T helper cell populations are often associated with dysbiosis, a poorly defined term describing alterations in the microbiota composition. Whilst these alterations in microbiota composition would once have been dismissed as an epiphenomenon, it is becoming increasingly apparent that compositional changes in the microbiota may both precede and play a causative role in the changes in immune cell function that drive these pathologies. In fact, it is now recognized that the immunomodulatory effects of dysbiosis are predominantly mediated through changes in the overall metabolic function of the microbiota resulting in an altered overall metabolite profile.11
While it is clear that the microbiota also interacts with other T cell subsets, such as intraepithelial lymphocytes (IEL),12 γδ-T cells,13,14 and MAIT cells,15,16 as well as non T cell populations,2 this review will focus on the intestinal CD4 T cell compartment. We will also highlight the impact of microbial CD4 T cell antigen mimicry in autoimmune disease and the role of the microbiota in cancer immune checkpoint blockade (ICB) therapy as examples of mechanistic (and immunological) links between intestinal CD4 T cells, microbes, and metabolites. Most of the current understanding has been obtained using gnotobiotic mouse models and mechanistic human data is sparse. Wherever possible, we will highlight the link to human data. Finally, we will also discuss open challenges in the field.
Differential induction of intestinal T helper cell responses by colonization with individual commensal bacteria or consortia
Colonization of germ-free mice with a diverse specific pathogen-free (SPF) microbiota results in the accumulation of CD4 T cells within the intestinal lamina propria.17,18 Under homeostatic conditions this occurs in the absence of overt inflammation indicating that this reflects a necessary immune adaption in response to intestinal colonization. The literature discussed here demonstrates that the nature of CD4 T cell subsets induced depends on what bacterial species are present and what metabolites are produced. These studies would not have been possible without the renaissance of experimental mouse gnotobiology, as precisely controlled in vivo experiments are essential to mechanistically study host-microbial interactions.19
T helper type 17 (Th17) cell induction by segmented filamentous bacteria (SFB)
The notion that different consortia of commensal bacteria or different individual bacterial species induce distinct intestinal T helper cell effector subtypes was initially sparked by the observation that mice obtained from various vendors displayed different intestinal T helper effector profiles, which correlated with differences in microbiota composition.20 Specifically, mice lacking segmented filamentous bacteria (SFB) displayed reduced Th17 levels in their intestines compared to genetically identical mice that were colonized with a microbiota that contained SFB.21,22 Using SFB monocolonized mice demonstrated that SFB was sufficient to induce Th17 cells, particularly in the small intestine (Fig. 1, Table 1). Induction of Th17 by SFB is dependent on MHCII expression by dendritic cells (DCs)23 and monocyte-derived macrophages,24 and these Th17 cells expressed a T cell receptor (TCR) specific for SFB antigens.23,25 Epithelial adhesion of SFB stimulates the production of serum amyloid A (SAA) and reactive oxygen species (ROS) by epithelial cells, important factors for Th17 cell differentiation.26,27 Th17 cells have pleiotropic functions and can act in a homeostatic or pro-inflammatory fashion and this balance was recently shown to be modulated by SAA proteins.28 In summary, epithelial adhesion of SFB triggers the transfer of T cell antigens into intestinal epithelial cells (IEC) which induces the activation of lamina propria SFB-specific Th17 cells.29 However, this mechanism does not seem to be a general feature of epithelium adherent bacteria. Hence, although epithelial adherence has been identified as an important characteristic for Th17-inducing bacteria, the downstream molecular mechanisms are likely to be microbe-specific.
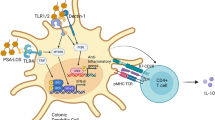
Microbial colonization has a profound impact on the intestinal CD4+ T cells compartment and provides signals leading to the differentiation of naïve T cells into Tbet+ Th1, GATA3+ Th2, RORγt+ Th17, FOXP3+ Treg, and BCL-6+ T follicular helper cells (TFH). From left to right: Bilophila wadsworthia a sulfite-reducing bacterium that flourishes on the sulfur contained in taurine induces a pro-inflammatory Th1 response. Ectopic colonization of oral Klebsiella species in the intestine induces a strong pro-inflammatory Th1 response. A consortium of 11 human commensals elicit intestinal Tbet+ Th1 and RORγt+ Th17 cells. Potent Th17 induction is also observed by epithelium adhering microbes such as Segmented filamentous bacteria (SFB), and Bifidobacterium adolescentis. A human gut consortium and a consortium of 20 strains isolated from a patient with ulcerative colitis can induce Th17 cells. During intestinal inflammation, Helicobacter species induce antigen-specific Th17 cells. Conversely, under homeostatic conditions Helicobacter induces Treg with specificity for the same epitopes. Bacteroides fragilis and Bifidobacterium bifidum promote the induction of Treg in a polysaccharide A (PSA) and cell surface β- glucan/galactan (CSGG) polysaccharide-dependent manner, respectively. Treg are further induced by Bifidobacterium infantis, Bacteroides thetaiotaomicron, Roseburia hominis, and Clostridium strains which are able to synthesize short chain fatty acids (SCFAs). Lactobacillus reuteri and Lactobacillus murinus metabolize tryptophan to indole which signals through aryl hydrocarbon receptor (AhR) in CD4+ intraepithelial lymphocytes (IEL) thereby contributing to the differentiation into CD4+CD8αα+ IELs. Akkermansia muciniphila induces an antigen-specific TFH response during homeostasis.
As SFB has not yet been identified in humans, a large set of individual commensal species derived from the human gut were screened for their ability to induce intestinal Th17 cells in mice.30 Bifidobacterium adolescentis was identified as one of several human symbiotic bacteria that elicited a robust intestinal Th17 population without provoking mucosal or systemic inflammation in monocolonized mice30 (Fig. 1, Table 1). Like SFB,26 B. adolescentis is also closely associated with the intestinal epithelium.
Regulatory T cell induction
Treg are crucial to maintain intestinal homeostasis by restraining inappropriate immune responses. It is generally accepted that there are two main subsets of Treg that express the FOXP3 transcription factor. These include thymus-derived natural Treg (nTreg), expressing Helios and Neuropilin-1, and peripherally induced Treg (pTreg), which are thought to be negative for Helios and Neuropilin-1.31 Microbially-induced pTreg can also express the transcription factor RORγt and these Treg have been shown to be particularly suppressive in intestinal inflammation.31 Importantly, the use of these markers is still controversial and is only useful in mouse models and not in humans.32,33
In addition to inducing Th17 cell differentiation, SFB colonization also induces Treg gene signatures in mice.21,34 This likely explains why SFB induction of Th17 cells does not result in overt inflammation. Due to their therapeutic potential, induction of Treg through modification of the microbiota composition and its metabolic function is a promising approach. A number of individual commensal species or consortia able to induce intestinal Treg response have been identified (Fig. 1, Table 1).
Treg induction by consortia
Initial studies identified several consortia of mouse or human-derived Clostridia species as potent inducers of intestinal Treg cells (Fig. 1, Table 1).35,36,37 Clostridia from clusters IV and XIVa collectively induced secretion of transforming growth factor (TGF)-β by intestinal epithelial cells, which drove the differentiation and expansion of pTreg cells in the colon.
Colonization with the murine altered Schaedler flora (ASF) also induced intestinal Treg responses (Table 1).38 In response to ASF colonization, control and regulation of Th1 and Th17 cells was mainly mediated by Treg production of IL-10, as Treg-deficiency or blocking the IL-10 receptor resulted in induction of Th1 and Th17 cells. Potent Treg induction in the colon was also observed following DSS treatment of ASF colonized mice, indicating an interplay between homeostatic and damage/inflammation-associated Treg induction.38 More recently, intestinal pTreg induction was also described following colonization with a slightly more complex stable defined moderately diverse murine microbiota (Table 1) (sDMDMm2/Oligo-MM12).39
Treg induction by individual species
Induction of intestinal Treg following colonization with microbial consortia raised the question whether individual commensal species alone can also promote this. It is important to realize that in vivo screening of individual commensal species that have the capacity to induce Treg (or any other T helper subtype) is limited by the inherent biology of commensal bacteria and bacterial consortia that often rely on cooperation to successfully colonize. For example, some obligate anaerobic species are not able to colonize a germ-free intestine, likely due to the increased oxygen levels, and require prior colonization of facultative anaerobes to reduce oxygen levels first.40 Alternatively, stable maintenance of a community structure likely relies on metabolic cooperation.41
Despite these limitations, an elegant study by Sefik et al. identified a number of individual human-derived commensal species with the potential to induce RORγ+ Treg following monocolonization.42 Clostridium ramosum and Bacteroides thetaiotaomicron were among the strongest inducers, consistent with the notion that certain Clostridia and Bacteroides species promote induction and maintenance of intestinal Treg (Fig. 1, Table 1). Another study found that pathobionts such as Helicobacter spp. induce antigen-specific Treg whose function is dependent on the transcription factor cMAF. Conversely, in disease-susceptible IL-10 deficient mice, Helicobacter spp. induced colitogenic Th17 cells with specificity for the same epitopes43 (Fig. 1, Table 1). In addition, Akkermansia muciniphila has also been shown to induce antigen-specific colonic Treg44 (Fig. 1, Table 1). Another prominent example for intestinal Treg induction, although less of a typical human commensal, is Bacteroides fragilis, which promotes Treg induction and IL-10 mediated anti-inflammatory responses from T cells and DCs in a toll-like receptor (TLR2)-dependent manner45,46,47 (Fig. 1, Table 1).
Monocolonization of germ-free mice with the human commensal Roseburia hominis also increased Treg in the lamina propria48 and the abundance of R. hominis is decreased in the fecal microbiota of patients with ulcerative colitis.49
A number of studies have performed large scale screens to mine the human gut microbiota for strains with immunomodulatory capacity (Table 1).50,51 However, the effect on the intestinal CD4 T compartment was not always precisely analyzed.
Importantly, the diet has a significant impact on the metabolism of individual commensal species and on the composition of consortia. Indeed, using antigen-free mice (germ-free being fed an elemental diet), it was shown that the vast majority of pTreg in the small intestine are induced by dietary antigens.52 Furthermore, a recent report demonstrated that the ketogenic diet causes a reduction in intestinal Th17 cells.53 The specific diet used in studies is therefore an important experimental factor to be considered. This is particularly important as mouse diets used by different research institutions are not always well defined or standardized. Therefore, careful experimental controls for the diet used should be included in any future studies.
T helper type 1 (Th1) cell induction
In conventionally housed mice with a complex undefined microbiota, it has been demonstrated that the microbiota limits Th1 and promotes Treg responses against intestinal antigens. This microbiota-induced immune tolerance was mediated by CX3CR1+ mononuclear phagocytes.54 Indeed, while a plethora of individual bacterial species with the capacity to induce intestinal Treg or Th17 cells have been identified, very few commensal species that promote a non-inflammatory homeostatic Th1 response have been described. The first study to identify Th1 inducers demonstrated that Klebsiella species, which normally colonize the oral cavity, can ectopically colonize the gut under dysbiotic conditions where they induce a strong Th1 response55 (Fig. 1, Table 1). This response was pro-inflammatory and appeared to be dependent on CD11b−CD103+ DCs and TLR signaling. More recently, a defined consortia of 11 bacterial strains from healthy humans that elicits interferon-γ secreting CD8+ T cells and anti-cancer immunity was identified.56 This study also reported induction of colonic Th1 and Th17 cells (Fig. 1, Table 1).
T follicular helper (TFH) cell induction
In Peyer’s Patches, TFH cells support affinity maturation of B cells in germinal centers and differentiation of B cells to IgA-secreting plasma cells.57 It has been demonstrated that microbially-derived extracellular ATP regulates TFH cells, which in turn regulates secretory IgA (SIgA) and microbial composition.58 A. muciniphila has been reported to induce antigen-specific TFH cells to promote systemic antigen-specific T-dependent IgA and IgG1 production. This finding indicates that commensal bacteria do not only modulate intestinal but also systemic adaptive immune responses during homeostasis59 (Fig. 1, Table 1).
The studies illustrate important milestones in the discovery of individual intestinal commensal species or consortia with the capacity to induce different T helper cell subsets. The literature covered here is by no means complete (Table 1) and we apologize to authors of studies that are not mentioned due to space restraints.
Metabolites involved in the induction of different intestinal T helper cell subsets
Although the immunological pathways and transcription factors required to induce and maintain the various T helper cell subsets are well described,60 the observation that different intestinal commensal species promote distinct intestinal T helper subsets raises the question of how microbial metabolites are involved in this process. While TGF-β is essential for the in vivo conversion of naïve CD4 T cells into pTreg,61 a number of microbial metabolites have been identified that further promote or enhance this process. So far, most studies have focused on the role of metabolites in Treg induction. However, recent studies are beginning to unravel the impact of metabolites on the induction of Th17 and Th1 cells.
Diet-derived metabolites
Although we will not focus specifically on the role of diet in this review, it is important to realize that the diet has a dramatic impact on the intestinal microbiota and hence on the production of microbial metabolites.62 Furthermore, it is difficult to categorize metabolites into purely dietary, microbial, or host derived as the original metabolite can be further modified the microbes or the host. Nevertheless, we will attempt to categorize the metabolites discussed here into certain categories.
Vitamins
Retinoic acid (RA) is a host metabolite derived from dietary vitamin A and produced by intestinal CD103+ DCs, IEC, and intestinal mesenchymal cells.63 RA enhances the differentiation of pTreg in the presence of TGF-β.64,65,66 Although RA promotes Treg induction and suppresses the development of Th17 cells,65 it is also required to elicit proinflammatory T helper cells in response to infection or mucosal vaccination.67 These findings suggest that RA functions in a context-dependent manner and is capable of mediating both anti- and pro-inflammatory effects. The vitamin B9 (folic acid), which is also derived from the diet or generated by members of the microbiota,68 was shown to be a survival factor for Treg that express high levels of folate receptor 4 (Fig. 2, Table 2).69,70 Furthermore, vitamin B3 (Niacin), a bacterial-derived product and GPR109a ligand, has been shown to act on DCs and favor the induction of Treg (Table 2).71 Importantly, as vitamins are by definition essential for many biochemical processes, studies using vitamin-deficient diets to study immune function are often difficult to interpret.
Adenosine triphosphate (ATP)
Luminal adenosine-5′-tri-phosphate (ATP) can be both host- as well as microbiota-derived and has been shown to promote the differentiation of intestinal Th17 cells72,73 (Table 2). Germ-free mice display greatly reduced numbers of lamina propria Th17 cells, which can be rescued by the administration of ATP.72 ATP induced the production of cytokines that favor Th17 differentiation in CD70highCD11clow DCs through P2X receptor signaling. Of note, SFB do not produce high levels of ATP, therefore, SFB-mediated Th17 cell induction likely occurs through a mechanism independent of ATP signaling.22
It has also been demonstrated that ATP released by bacteria limits TFH cell abundance in Peyer’s Patches via the ATP-gated ionotropic P2X7 receptor, which results in decreased production of T-dependent SIgA in the small intestine.58 Increased SIgA can lead to changes in composition of the commensal bacteria, which has been shown to affect glucose metabolism and fat deposition in mice.58 Conversely, blocking microbial production of ATP can also increase specific IgA responses to live and inactivated oral vaccines.74 These studies illustrate that modulation of ATP in the small intestine can affect high-affinity IgA responses against gut colonizing bacteria.
Short chain fatty acids (SCFA)
The SCFA acetate, butyrate and propionate are generated by microbial fermentation of dietary fibers. SCFA are cardinal examples of metabolites with an immuno-modulatory effect on the mucosal CD4 T cell compartment (Fig. 2, Table 2). Multiple bacterial groups are able to produce acetate,11,39 and Clostridia species belonging to cluster IV and XIVa are prominent sources of butyrate in the gut.11 Propionate is produced by species of the phylum Bacteroides and Firmicutes.11 Acetate and butyrate, in particular, have been shown to promote intestinal Treg induction and function through multiple mechanisms.75,76,77 These include the upregulation of Foxp3 expression in CD4 T cells by enhanced Histone 3 acetylation of promoter and conserved non-coding sequence regions by histone deacetylase (HDAC) inhibition.76,77 Moreover, the effects of SCFA are mediated by binding to free fatty acid receptors such as G-protein coupled receptors (GPCRs) expressed on CD4 T cells (GPR43, also known as FAAR2) and DCs (GPR109A).71,75 Furthermore, SCFA upregulate GPR15 expression leading to Treg accumulation in the colon.75 More recently, it has been shown that SCFA promote T cell differentiation into both effector and regulatory T cells in a context-dependent manner. Th1 and Th17 cell differentiation was mediated by the HDAC inhibitory activity of SCFA in T cells, which subsequently resulted in enhanced mTOR-S6K activity required for differentiation and cytokine expression.78
In contrast to SCFA, dietary long chain-fatty acids enhance differentiation and proliferation of Th1 and Th17 cells.79 Furthermore, dietary fats derived from a high-fat diet induced changes in the ileum microbiota composition leading to a reduction in intestinal Th17 cells.80
Several groups have shown a protective effect of SCFA in mouse models of colitis.75,77 Similarly, in IBD patients microbial dysbiosis was associated with a decrease of SCFA producing bacteria and corresponding lower levels of fecal SCFA.11,49,81,82 Furthermore, fecal microbiota transplantation (FMT) from healthy donors into pediatric ulcerative colitis patients resulted in increased diversity, abundance of Clostridia, and fecal butyrate levels.83 These studies indicate that SCFA also play an important functional role in humans.
Microbial polysaccharides
Polysaccharide A (PSA) produced by B. fragilis is a good example of a bacterial compound with immuno-modulatory effects84 (Figs. 1 and 2, Table 2). PSA was reported to protect mice from experimental colitis.85 This protective effect was mediated by IL-10 production by intestinal Treg, in a TLR2-dependent mechanism,46 and also via indirect effects on conventional DCs47 or plasmacytoid DCs.86 Furthermore, it was shown that B. fragilis releases PSA via outer-membrane vesicles (OMV) which are delivered to DCs.47,87 These OMV-primed DCs induce intestinal Treg and thereby protect animals from experimental colitis. Similarly, cell surface β-glucan/galactan (CSGG) polysaccharides of Bifidobacterium bifidum were shown to induce intestinal Treg capable of suppressing intestinal inflammation via activation of DCs in a TLR2-dependent manner88 (Fig. 1, Table 2).
Aryl hydrocarbon receptor (AhR) ligands
Generally, AhR is required for the development and homeostasis of various cell types, including γδIELs,89 Th17, and Treg, with an important role in maintaining host-microbiota mutualism. Signaling through AhR has been shown to regulate Treg and Th17 cell differentiation in a ligand-specific manner.90,91,92 Recent work revealed that the AhR-Foxp3-RORγt axis controls gut homing of CD4 T cells by regulating the expression of GPR15.93
The microbiota can modulate the immune system through activation of the AhR transcription factor. AhR ligands can be derived from many sources. Besides xenobiotic compounds, such as dioxin, AhR also recognizes natural ligands derived from cruciferous vegetables, as well as ligands produced by the microbiota through tryptophan metabolism.94 For example, Lactobacillus reuteri provides indole derivates, such as indole-3 lactic acid, of dietary tryptophan that activate AhR. This induces intraepithelial CD4+CD8αα+ T cells via downregulation of the transcription factor ThPOK (Fig. 1, Table 2).12
Catabolism of tryptophan into the AhR ligand kynurenine, mediated by indoleamine-2,3-dioxygenase (IDO) activity in DCs and IEC, favors pTreg differentiation and controls effector T cell activity.95,96,97 Interestingly, IEC of mice colonized with Clostridium express high level of IDO.36
Furthermore, the importance of microbial-derived AhR ligands in intestinal immune education was recently highlighted using a model of gestational colonization.98 This study revealed that AhR ligands, produced by the maternal microbiota, were transferred to the offspring postnatally through the milk and shaped the offspring’s intestinal innate lymphoid cells (ILC) 3 population.
Bile acids (BA)
BA are small molecules that are synthesized from cholesterol in the liver and are further metabolized by the intestinal microbiota, including deconjugation of glycine or taurine and biotransformation of unconjugated primary BA to secondary BA. Hence, in comparison to conventionally housed animals, the BA profile of germ-free animals is markedly different with a reduced diversity and overall abundance of secondary BA.99 BA function as signaling molecules with pleiotropic metabolic and immune effects through dynamic interactions with host receptors and the microbiota resulting in a complex interplay between BA, the microbiota and the host immune system. For example, mice fed with a high-fat diet displayed an altered composition of conjugated BA, resulting in an expansion of Bilophila wadsworthia, a sulfite-reducing bacterium100 (Fig. 1). These changes were associated with enhanced intestinal Th1 responses and the development of colitis in genetically susceptible IL-10-deficient animals. Furthermore, a study by Cao et al. showed that BA induce T cell mediated inflammation in the ileum, unless T cells are protected by the xenobiotic transporter Mdr1.101
Two recent studies have identified additional anti-inflammatory roles for BA metabolites that modulate intestinal CD4 T cell responses. Song et al. demonstrate the importance of microbial BA in the development of colonic RORγt+ Treg in mice102 (Table 2). Genetic ablation of BA metabolizing genes in individual symbiotic bacteria led to a reduction of Th17 cells in vivo. Conversely, restoration of the BA pool enhanced colonic RORγt+ Treg and ameliorated gut inflammation through the BA/vitamin D receptor axis (Fig. 3). Moreover, Hang et al. identified two distinct derivates of lithocholic acid (LCA) as T cell regulators in mice.103 While 3-oxoLCA inhibited the differentiation of Th17 cells by binding to RORγt and inhibiting its transcriptional activity, isoalloLCA enhanced Treg differentiation (Table 2). Collectively, these studies revealed novel mechanisms that regulate CD4 T cell responses through BA metabolites.
In human inflammatory bowel disease (IBD), altered fecal BA profiles characterized by an increase in conjugated primary BA and a decrease in secondary BA levels have been described.104 Furthermore, reduced levels of stool LCA and deoxycholic acid (DCA) were reported in colectomy treated patients with ulcerative colitis compared to familial adenomatous polyposis pouch patients.105 In this study, diminished secondary BA were linked with altered microbial composition and lower expression of genes required for the conversion of primary to secondary BA. In animal models of acute and chronic colitis, supplementation with LCA and DCA reduced intestinal inflammation. Of note, LCA has been demonstrated to impair Th1 activation, evidenced by IFNγ and TNFα production, through vitamin D receptor signaling.106
Collectively, these studies demonstrate that the intestinal CD4 T cell compartment can be activated by diverse microbial taxa and a variety of different mechanisms. High degrees of redundancy in these processes are crucial for the control and the maintenance of intestinal homeostasis. Of note, some of these microbes that elicit tolerogenic immune adaptations have also been implicated in disease pathogenesis in experimental models of inflammation.107,108,109 This finding suggests that these beneficial effects are context-dependent and modulated by host-genetics factors.
The wide-spread use of targeted as well as untargeted metabolomics methodology in microbiome research will undoubtedly result in the identification of additional microbial metabolites involved in microbiota-immune crosstalk.
Intestinal CD4 T helper cells in disease and therapy
The role of intestinal T helper cells in disease is an extremely large topic,60 particularly when also considering the role of metabolites in disease. It is well established that intestinal T helper cells as well as metabolites are involved in intestinal disorders, which has been reviewed in detail elsewhere.11,110,111 Some mouse models for systemic autoimmune disorders, such as experimental autoimmune encephalomyelitis (EAE), have also demonstrated a potential link to intestinal CD4 T helper cells. However, the precise immunological mechanisms as well as the role of TCR reactivity remained unclear.109 Therefore, we will highlight recent work that elucidated the role of intestinal CD4 T helper cells in systemic (non-intestinal) diseases where the immunological mechanism involved has been identified to be microbial antigen mimicry. First, we will provide a short summary on what is known about antigen-specificity of intestinal T helper cells, which has been previously reviewed in detail.112
Antigen-specificity of intestinal T helper cell responses
Most of the current literature on the role of metabolites on intestinal T helper cell responses did not address whether the observed effects were antigen-specific or not. This is likely due to the fact that the role of antigen-specificity remains difficult to address experimentally. However, antigen-specificity is an important aspect, especially in the context of microbiota-autoimmune crosstalk, which includes microbial antigen mimicry as discussed later.
The precise requirements for antigen-specificity for induction and effector function of intestinal regulatory T cells remains unclear. When considering antigen-specificity of the intestinal Treg compartment both peripherally induced pTreg and thymus-derived natural nTreg populations needs to be taken into consideration.36,38,113 nTreg have a repertoire skewed towards self-reactivity, while pTreg have a repertoire similar to that of naïve CD4 T cells.114,115 Although elegant studies have cloned the TCR of intestinal Treg and identified potential bacterial candidates as the source of the antigen, the precise antigen itself was not identified.116,117 As mentioned above, Helicobacter induce antigen-specific Treg43 and the antigen-specificity of Th17 cells induced by SFB has been carefully characterized.23,25,29 Furthermore, A. muciniphila induces antigen-specific TFH and Treg cells.44,59
The experimental difficulties generated by the combination of a complex microbiota and the presence of a diverse TCR repertoire resulted in the establishment of several simplified experimental models that facilitate the study of antigen-specificity in host-microbiota crosstalk (Table 3). These models combine cloning of TCRs, use of transgenic TCRs with known specificity, and use of defined gnotobiotic mouse models to generate simplified experimental conditions. These tools have been instrumental in gaining valuable insight on the role of antigen-specificity of intestinal T helper cells.
Intestinal commensal-specific CD4 T cells in humans are even harder to investigate and most studies have been performed in the context of inflammatory condition using blood samples, which does not reflect true intestinal CD4 T cells.118,119,120,121 One elegant study did demonstrate however that microbiota-specific CD4 T cells are abundant in the circulation of healthy individuals.122
Antigen mimicry and autoimmune disease
A large number of studies linking microbiota, intestinal T helper cells, and disease are correlative in nature and hence lack a detailed molecular or immunological mechanism.123 We will focus here on recent literature implicating molecular mimicry of microbial and host CD4 T cell epitopes in autoimmune diseases. Molecular mimicry of cross-reactive antigenic epitopes between microbes and host self-peptides has been thought of as a potential mechanism to drive activation of auto-reactive T cells.124,125 In addition to the previously mentioned EAE study,109 the microbiota and CD4 T cell responses have also been implicated in mouse models of arthritis.126,127 While no specific cross-reactive microbial antigen mimic has yet been identified in arthritis, it has been shown that intestinal Th17 cells expressing dual TCRs are involved in lung autoimmunity, which is a major cause of arthritis-related mortality.128 While this is not classical molecular antigen mimicry, it was one of the first studies reporting the involvement of TCR specificity of intestinal CD4 T cells in systemic autoimmunity. Microbiota-dependent activation of an autoreactive TCR was also involved in the pathogenesis of autoimmune uveitis.129,130,131 In this study, the activation of retina-specific T cells involved signaling through the autoreactive TCR induced by a microbial antigen and was independent of the endogenous retinal autoantigen.129 However, the precise bacterial species and antigen involved have not been identified. Furthermore, skin, oral, and gut commensal orthologs of the human lupus autoantigen Ro60 were able to activate Ro60-specific CD4 memory T cell clones from lupus patients, implicating T cell cross-reactivity and microbial antigen mimicry in lupus.132 A recent study provided proof of the existence of a specific microbial peptide mimic within β-galactosidase expressed by B. thetaiotaomicron. This molecular mimic induced autoimmune T helper cells that also recognize the autoantigen myosin heavy chain 6 (MYH6), thus causing cardiomyopathy.133 This study also linked their findings to a human patient population. Microbial antigen mimicry in autoimmune disease has not only been identified for CD4 T cells but also for CD8 T cells in Type 1 diabetes, for example.134 It is also important to note that microbial mimicry of host autoantigens does not necessarily have to be detrimental but can also be protective, as demonstrated in a mouse model of colitis.135 Taken together, these studies clearly demonstrate microbial antigen mimicry as an important mechanism by which intestinal CD4 T helper cell responses can either promote or protect from systemic autoimmune disease. It will be interesting to study whether bacterial or host metabolites are involved in determining whether antigen mimicry is detrimental or beneficial in an autoimmune setting.
Microbiota-mediated T helper cell responses in cancer ICB immunotherapy
We are using ICB as an example for how commensal bacteria-mediated intestinal CD4 T cell responses can be used to promote disease therapy. The most widely used current ICB strategies include monoclonal antibodies targeting cytotoxic T lymphocyte-associated protein 4 (CTLA-4) and/or programmed cell death protein (PD-1) as well as its ligand PD-L1. Seminal work using murine tumor models demonstrated that the gut microbiota influences the efficacy of ICB therapy136,137 and more recent work translated these findings to cancer patients.56,138,139,140 For example, enrichment of A. muciniphila in the fecal microbiota of patients with epithelial cancer correlated with a favorable clinical outcome following ICB therapy.138 Furthermore, a study involving patients with metastatic melanoma receiving ICB therapy observed an increased abundance of eight microbial species including Bifidobacterium longum in ICB responders.140 Another study on ICB therapy in metastatic melanoma patients found a significant enrichment of the Faecalibacterium genus in responders, whereas non-responders displayed a higher abundance of the Bacteroidales order.139
Beyond these correlative patient data, it has been shown that germ-free or antibiotic treated tumor-bearing mice receiving fecal microbiota transplantation (FMT) from patients responding to ICB improved the response to anti-PD-1 treatment.138,139,140 In contrast, mice receiving FMT from non-responders showed poor response to ICB therapy. Of note, oral gavage of A. muciniphila following FMT with non-responder’s feces restored efficacy of ICB therapy in vivo, suggesting that primary resistance to ICB therapy may partially be due to the absence of specific bacteria.138 The beneficial effect of A. muciniphila in tumor-bearing mice was dependent on increased secretion of IL-12 by DCs promoting the accumulation of CCR9+CXCR3+CD4+ T cells in lymph nodes and tumor.138 Similarly, B. fragilis-mediated Th1 immune responses following anti-CTLA-4 treatment were also IL-12 dependent.137
Collectively, these studies provide compelling evidence that the intestinal microbiota influences anti-tumor immunity. Although the specific microbes associated with enhanced clinical response vary between reports, it is likely that various members of the human microbiota can modulate the response to ICB therapy. A study by Tanoue et al. revealed that a consortium of 11 bacteria isolated from the human gut microbiota induced IFNγ-producing CD8 T cells and, to a lower extent, also CD4 T cells. These bacteria-induced anti-tumor immunity by itself and further promoted the efficacy of ICB inhibitors in multiple tumor models.56 The authors suggested that circulating metabolites produced or induced by these microbes might modulate systemic T cells. Indeed, several immunomodulatory metabolites, such as mevalonate and dimethylglycine, were elevated in the caecal contents and serum of gnotobiotic mice colonized with the 11 strain consortium. The challenge in this field is now to move on from identifying bacteria that enhance ICB therapy towards molecular mechanisms of how these bacteria improve ICB-therapy efficacy.
Conclusion
The various topics discussed in this review are summarized in Fig. 3. While understanding microbial composition, diversity, and richness are already complex enough, it is now clear that a functional understanding of the microbiota requires a more in-depth understanding of the immunomodulatory capacity of microbial metabolites. This added complexity poses both experimental and analytical challenges. One important experimental consideration is the impact of differing diets on microbiota composition and metabolic activity. It is therefore important to continue to experimentally dissect the interplay between microbes, metabolites, diet, immune system, and the host to study host-microbial crosstalk and how this modulates the dynamics of intestinal T helper cell phenotypes and function. Continuing development of computational analysis tools for the analysis of metabolomic data, similar to the tools available for microbiota composition analysis, is important to overcome analytical challenges non-specialist labs currently face.141 Another important challenge is the translation from mouse models to humans. In vitro screening methods for human-derived commensal microbes are key for translating mouse data to the human situation.142 Furthermore, recent studies in wild mice that harbor a wild microbiota, which is considerably more diverse than standard SPF laboratory mice, have demonstrated immune phenotypes that more closely resemble the human situation.143 Disease phenotypes and therapies in these wild mice also better reflected what is observed in human patients.144 Therefore, while defined experimental germ-free and gnotobiotic mouse models are essential to identify molecular disease mechanisms together with specific microbial species and metabolites involved in a given disease model, wild mice might provide an important tool to translate these mechanistic findings into an experimental in vivo model that more closely resembles the human situation.
Charles Janeway famously referred to the use of adjuvants by experimental immunologist as their “dirty little secret”, which lead to the formulation of his hypothesis of the existence of evolutionary conserved pattern recognition receptors.145 Wild mice with a very complex and undefined microbiota may hold their own “dirty little secret”, which will help us to better translate disease mechanisms identified in defined mouse models to the human situation.
References
Geuking, M. Co-evolution of microbes and immunity and its consequences for modern-day life. In: The Human Microbiota and Chronic Disease: Dysbiosis as a Cause of Human Patholog (ed. Luigi N. B. H.) pp 155–164 (John Wiley & Sons, Inc, 2016).
Belkaid, Y. & Harrison, O. J. Homeostatic Immunity and the Microbiota. Immunity 46, 562–576 (2017).
van Tilburg Bernardes, E. et al. Intestinal fungi are causally implicated in microbiome assembly and immune development in mice. Nat. Commun. 11, 2577 (2020).
Bacher, P. et al. Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against Candida albicans. Cell 176, 1340–1355. e1315 (2019).
Chiaro, T. R. et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci. Transl. Med. 9, eaaf9044 (2017).
Iliev, I. D. et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317 (2012).
Limon, J. J. et al. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe 25, 377–388. e376 (2019).
Wikoff, W. R. et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl Acad. Sci. USA 106, 3698–3703 (2009).
Claus, S. P. et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol. Syst. Biol. 4, 219 (2008).
Uchimura, Y. et al. Antibodies set boundaries limiting microbial metabolite penetration and the resultant mammalian host response. Immunity 49, 545–559 e545 (2018).
Lavelle, A. & Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 17, 223–237 (2020).
Cervantes-Barragan, L. et al. Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science 357, 806–810 (2017).
Hagihara, M. et al. Clostridium butyricum modulates the microbiome to protect intestinal barrier function in mice with antibiotic-induced dysbiosis. iScience 23, 100772 (2020).
Duan, J., Chung, H., Troy, E. & Kasper, D. L. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host Microbe 7, 140–150 (2010).
Treiner, E. et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003).
Constantinides, M. G. et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366, eaax6624 (2019).
Macpherson, A. J. & Harris, N. L. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4, 478–485 (2004).
Macpherson, A. J., Martinic, M. M. & Harris, N. The functions of mucosal T cells in containing the indigenous commensal flora of the intestine. Cell Mol. Life Sci. 59, 2088–2096 (2002).
McCoy, K. D., Geuking, M. B. & Ronchi, F. Gut microbiome standardization in control and experimental mice. Curr. Protoc. Immunol. 117, 23 21 21–23 21 13 (2017).
Ivanov, I. I. et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008).
Gaboriau-Routhiau, V. et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Goto, Y. et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40, 594–607 (2014).
Panea, C. et al. Intestinal monocyte-derived macrophages control commensal-specific Th17 responses. Cell Rep. 12, 1314–1324 (2015).
Yang, Y. et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156 (2014).
Atarashi, K. et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380 (2015).
Sano, T. et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 163, 381–393 (2015).
Lee, J. Y. et al. Serum amyloid A proteins induce pathogenic Th17 cells and promote inflammatory disease. Cell 180, 79–91. e16 (2020).
Ladinsky, M. S. et al. Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science 363, eaat4042 (2019).
Tan, T. G. et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl Acad. Sci. USA 113, E8141–E8150 (2016).
Tanoue, T., Atarashi, K. & Honda, K. Development and maintenance of intestinal regulatory T cells. Nat. Rev. Immunol. 16, 295–309 (2016).
Bruder, D. et al. Neuropilin-1: a surface marker of regulatory T cells. Eur. J. Immunol. 34, 623–630 (2004).
Milpied, P. et al. Neuropilin-1 is not a marker of human Foxp3+ Treg. Eur. J. Immunol. 39, 1466–1471 (2009).
Ohnmacht, C. et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 349, 989–993 (2015).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013).
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011).
Narushima, S. et al. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut Microbes 5, 333–339 (2014).
Geuking, M. B. et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34, 794–806 (2011).
Wyss, M. et al. Using precisely defined in vivo microbiotas to understand microbial regulation of IgE. Front Immunol. 10, 3107 (2019).
Becker, N., Kunath, J., Loh, G. & Blaut, M. Human intestinal microbiota: characterization of a simplified and stable gnotobiotic rat model. Gut Microbes 2, 25–33 (2011).
Coyte, K. Z. & Rakoff-Nahoum, S. Understanding competition and cooperation within the mammalian gut microbiome. Curr. Biol. 29, R538–R544 (2019).
Sefik, E. et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 349, 993–997 (2015).
Xu, M. et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018).
Kuczma, M. P. et al. Commensal epitopes drive differentiation of colonic Tregs. Sci. Adv. 6, eaaz3186 (2020).
Round, J. L. et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977 (2011).
Round, J. L. & Mazmanian, S. K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl Acad. Sci. USA 107, 12204–12209 (2010).
Shen, Y. et al. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12, 509–520 (2012).
Patterson, A. M. et al. Human gut symbiont roseburia hominis promotes and regulates innate immunity. Front Immunol. 8, 1166 (2017).
Machiels, K. et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275–1283 (2014).
Geva-Zatorsky, N. et al. Mining the human gut microbiota for immunomodulatory organisms. Cell 168, 928–943. e911 (2017).
Faith, J. J., Ahern, P. P., Ridaura, V. K., Cheng, J. & Gordon, J. I. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci. Transl. Med. 6, 220ra211 (2014).
Kim, K. S. et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 351, 858–863 (2016).
Ang, Q. Y. et al. Ketogenic diets alter the gut microbiome resulting in decreased intestinal Th17 cells. Cell 181, 1263–1275 (2020).
Kim, M. et al. Critical role for the microbiota in CX3CR1(+) intestinal mononuclear phagocyte regulation of intestinal T cell responses. Immunity 49, 151–163 e155 (2018).
Atarashi, K. et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 358, 359–365 (2017).
Tanoue, T. et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605 (2019).
Crotty, S. Follicular helper CD4 T cells (TFH). Annu Rev. Immunol. 29, 621–663 (2011).
Perruzza, L. et al. T follicular helper cells promote a beneficial gut ecosystem for host metabolic homeostasis by sensing microbiota-derived extracellular ATP. Cell Rep. 18, 2566–2575 (2017).
Ansaldo, E. et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184 (2019).
Brown, E. M., Kenny, D. J. & Xavier, R. J. Gut microbiota regulation of T cells during inflammation and autoimmunity. Annu Rev. Immunol. 37, 599–624 (2019).
Chen, W. et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886 (2003).
Zmora, N., Suez, J. & Elinav, E. You are what you eat: diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 16, 35–56 (2019).
Thomson, C. A., Nibbs, R. J., McCoy, K. D. & Mowat, A. M. Immunological roles of intestinal mesenchymal cells. Immunology 160, 313–324 (2020).
Coombes, J. L. et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 204, 1757–1764 (2007).
Mucida, D. et al. Retinoic acid can directly promote TGF-beta-mediated Foxp3(+) Treg cell conversion of naive T cells. Immunity 30, 471–472 (2009). Author reply 472-473.
Sun, C. M. et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 204, 1775–1785 (2007).
Hall, J. A. et al. Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity 34, 435–447 (2011).
Yoshii, K., Hosomi, K., Sawane, K. & Kunisawa, J. Metabolism of dietary and microbial vitamin B family in the regulation of host immunity. Front Nutr. 6, 48 (2019).
Kinoshita, M. et al. Dietary folic acid promotes survival of Foxp3+ regulatory T cells in the colon. J. Immunol. 189, 2869–2878 (2012).
Kunisawa, J., Hashimoto, E., Ishikawa, I. & Kiyono, H. A pivotal role of vitamin B9 in the maintenance of regulatory T cells in vitro and in vivo. PLoS ONE 7, e32094 (2012).
Singh, N. et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139 (2014).
Atarashi, K. et al. ATP drives lamina propria T(H)17 cell differentiation. Nature 455, 808–812 (2008).
Kusu, T. et al. Ecto-nucleoside triphosphate diphosphohydrolase 7 controls Th17 cell responses through regulation of luminal ATP in the small intestine. J. Immunol. 190, 774–783 (2013).
Proietti, M. et al. ATP released by intestinal bacteria limits the generation of protective IgA against enteropathogens. Nat. Commun. 10, 250 (2019).
Smith, P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Park, J. et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 8, 80–93 (2015).
Haghikia, A. et al. Dietary fatty acids directly impact central nervous system autoimmunity via the small intestine. Immunity 43, 817–829 (2015).
Garidou, L. et al. The gut microbiota regulates intestinal CD4 T cells expressing rorgammat and controls metabolic disease. Cell Metab. 22, 100–112 (2015).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
Nusbaum, D. J. et al. Gut microbial and metabolomic profiles after fecal microbiota transplantation in pediatric ulcerative colitis patients. FEMS Microbiol. Ecol. 94, fiy133 (2018).
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O. & Kasper, D. L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005).
Mazmanian, S. K., Round, J. L. & Kasper, D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008).
Dasgupta, S., Erturk-Hasdemir, D., Ochoa-Reparaz, J., Reinecker, H. C. & Kasper, D. L. Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe 15, 413–423 (2014).
Chu, H. et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120 (2016).
Verma, R. et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3(+) regulatory T cells. Sci. Immunol. 3, eaat6975 (2018).
Li, Y. et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147, 629–640 (2011).
Quintana, F. J. et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71 (2008).
Veldhoen, M. et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453, 106–109 (2008).
Kimura, A., Naka, T., Nohara, K., Fujii-Kuriyama, Y. & Kishimoto, T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc. Natl Acad. Sci. USA 105, 9721–9726 (2008).
Xiong, L. et al. Ahr-Foxp3-RORgammat axis controls gut homing of CD4(+) T cells by regulating GPR15. Sci. Immunol. 5, eaaz7277 (2020).
Shinde, R. & McGaha, T. L. The Aryl hydrocarbon receptor: connecting immunity to the microenvironment. Trends Immunol. 39, 1005–1020 (2018).
Mezrich, J. D. et al. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 185, 3190–3198 (2010).
Munn, D. H. & Mellor, A. L. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 34, 137–143 (2013).
Matteoli, G. et al. Gut CD103+ dendritic cells express indoleamine 2,3-dioxygenase which influences T regulatory/T effector cell balance and oral tolerance induction. Gut 59, 595–604 (2010).
Gomez de Aguero, M. et al. The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 (2016).
Sayin, S. I. et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 (2013).
Devkota, S. et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 487, 104–108 (2012).
Cao, W. et al. The xenobiotic transporter Mdr1 enforces T cell homeostasis in the presence of intestinal bile acids. Immunity 47, 1182–1196 e1110 (2017).
Song, X. et al. Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature 577, 410–415 (2020).
Hang, S. et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576, 143–148 (2019).
Duboc, H. et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 62, 531–539 (2013).
Sinha, S. R. et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe 27, 659–670 e655 (2020).
Pols, T. W. H. et al. Lithocholic acid controls adaptive immune responses by inhibition of Th1 activation through the Vitamin D receptor. PLoS ONE 12, e0176715 (2017).
Bloom, S. M. et al. Commensal bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe 9, 390–403 (2011).
Stepankova, R. et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm. Bowel Dis. 13, 1202–1211 (2007).
Lee, Y. K., Menezes, J. S., Umesaki, Y. & Mazmanian, S. K. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl Acad. Sci. USA 108, 4615–4622 (2011).
Skelly, A. N., Sato, Y., Kearney, S. & Honda, K. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat. Rev. Immunol. 19, 305–323 (2019).
Imam, T., Park, S., Kaplan, M. H. & Olson, M. R. Effector T helper cell subsets in inflammatory bowel diseases. Front Immunol. 9, 1212 (2018).
Russler-Germain, E. V., Rengarajan, S. & Hsieh, C. S. Antigen-specific regulatory T-cell responses to intestinal microbiota. Mucosal Immunol. 10, 1375–1386 (2017).
Cebula, A. et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 497, 258–262 (2013).
Curotto de Lafaille, M. A. & Lafaille, J. J. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity 30, 626–635 (2009).
Savage, P. A., Klawon, D. E. J. & Miller, C. H. Regulatory T cell development. Annu Rev. Immunol. 38, 421–453 (2020).
Lathrop, S. K. et al. Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254 (2011).
Lathrop, S. K., Santacruz, N. A., Pham, D., Luo, J. & Hsieh, C. S. Antigen-specific peripheral shaping of the natural regulatory T cell population. J. Exp. Med. 205, 3105–3117 (2008).
Calderon-Gomez, E. et al. Commensal-specific CD4(+) cells from patients with crohn’s disease have a T-helper 17 inflammatory profile. Gastroenterology 151, 489–500 e483 (2016).
Ergin, A. et al. Impaired peripheral Th1 CD4+ T cell response to Escherichia coli proteins in patients with Crohn’s disease and ankylosing spondylitis. J. Clin. Immunol. 31, 998–1009 (2011).
Shen, C., Landers, C. J., Derkowski, C., Elson, C. O. & Targan, S. R. Enhanced CBir1-specific innate and adaptive immune responses in Crohn’s disease. Inflamm. Bowel Dis. 14, 1641–1651 (2008).
Sorini, C., Cardoso, R. F., Gagliani, N. & Villablanca, E. J. Commensal bacteria-specific CD4(+) T cell responses in health and disease. Front Immunol. 9, 2667 (2018).
Hegazy, A. N. et al. Circulating and tissue-resident CD4(+) T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology 153, 1320–1337. e1316 (2017).
Walter, J., Armet, A. M., Finlay, B. B. & Shanahan, F. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell 180, 221–232 (2020).
Albert, L. J. & Inman, R. D. Molecular mimicry and autoimmunity. N. Engl. J. Med. 341, 2068–2074 (1999).
Munz, C., Lunemann, J. D., Getts, M. T. & Miller, S. D. Antiviral immune responses: triggers of or triggered by autoimmunity? Nat. Rev. Immunol. 9, 246–258 (2009).
Wu, H. J. et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010).
Teng, F. et al. Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of Peyer’s patch T follicular helper cells. Immunity 44, 875–888 (2016).
Bradley, C. P. et al. Segmented filamentous bacteria provoke lung autoimmunity by inducing gut-lung axis Th17 cells expressing dual TCRs. Cell Host Microbe 22, 697–704 e694 (2017).
Horai, R. et al. Microbiota-dependent activation of an autoreactive T cell receptor provokes autoimmunity in an immunologically privileged site. Immunity 43, 343–353 (2015).
Horai, R. & Caspi, R. R. Microbiome and autoimmune uveitis. Front Immunol. 10, 232 (2019).
Zarate-Blades, C. R. et al. Gut microbiota as a source of a surrogate antigen that triggers autoimmunity in an immune privileged site. Gut Microbes 8, 59–66 (2017).
Greiling, T. M. et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transl. Med. 10, eaan2306 (2018).
Gil-Cruz, C. et al. Microbiota-derived peptide mimics drive lethal inflammatory cardiomyopathy. Science 366, 881–886 (2019).
Tai, N. et al. Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J. Exp. Med. 213, 2129–2146 (2016).
Hebbandi Nanjundappa, R. et al. A gut microbial mimic that hijacks diabetogenic autoreactivity to suppress colitis. Cell 171, 655–667 e617 (2017).
Sivan, A. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015).
Vetizou, M. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Matson, V. et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018).
Ivanov, I. Gut-host crosstalk: methodological and computational challenges. Dig. Dis. Sci. 65, 686–694 (2020).
Geiger, R., Duhen, T., Lanzavecchia, A. & Sallusto, F. Human naive and memory CD4+ T cell repertoires specific for naturally processed antigens analyzed using libraries of amplified T cells. J. Exp. Med. 206, 1525–1534 (2009).
Beura, L. K. et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516 (2016).
Rosshart, S. P. et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365, eaaw4361 (2019).
Janeway, C. A. Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989).
Jeon, S. G. et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 8, e1002714 (2012).
Konieczna, P. et al. Immunomodulation by Bifidobacterium infantis 35624 in the murine lamina propria requires retinoic acid-dependent and independent mechanisms. PLoS ONE 8, e62617 (2013).
Kashiwagi, I. et al. Smad2 and Smad3 inversely regulate TGF-beta autoinduction in clostridium butyricum-activated dendritic cells. Immunity 43, 65–79 (2015).
Hayashi, A. et al. A single strain of Clostridium butyricum induces intestinal IL-10-producing macrophages to suppress acute experimental colitis in mice. Cell Host Microbe 13, 711–722 (2013).
Li, Y. N. et al. Effect of oral feeding with Clostridium leptum on regulatory T-cell responses and allergic airway inflammation in mice. Ann. Allergy Asthma Immunol. 109, 201–207 (2012).
Sarrabayrouse, G. et al. CD4CD8alphaalpha lymphocytes, a novel human regulatory T cell subset induced by colonic bacteria and deficient in patients with inflammatory bowel disease. PLoS Biol. 12, e1001833 (2014).
Tang, C. et al. Inhibition of dectin-1 signaling ameliorates colitis by inducing lactobacillus-mediated regulatory T cell expansion in the intestine. Cell Host Microbe 18, 183–197 (2015).
Karimi, K., Inman, M. D., Bienenstock, J. & Forsythe, P. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am. J. Respir. Crit. Care Med. 179, 186–193 (2009).
Jang, S. O. et al. Asthma prevention by lactobacillus rhamnosus in a mouse model is associated with CD4(+)CD25(+)Foxp3(+) T cells. Allergy Asthma Immunol. Res 4, 150–156 (2012).
Nakamoto, N. et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat. Microbiol 4, 492–503 (2019).
Wu, W. et al. Commensal A4 bacteria inhibit intestinal Th2-cell responses through induction of dendritic cell TGF-beta production. Eur. J. Immunol. 46, 1162–1167 (2016).
Lavasani, S. et al. A novel probiotic mixture exerts a therapeutic effect on experimental autoimmune encephalomyelitis mediated by IL-10 producing regulatory T cells. PLoS ONE 5, e9009 (2010).
Calcinaro, F. et al. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia 48, 1565–1575 (2005).
Di Giacinto, C., Marinaro, M., Sanchez, M., Strober, W. & Boirivant, M. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-beta-bearing regulatory cells. J. Immunol. 174, 3237–3246 (2005).
Cong, Y., Feng, T., Fujihashi, K., Schoeb, T. R. & Elson, C. O. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc. Natl Acad. Sci. USA 106, 19256–19261 (2009).
Kwong Chung, C. K., Ronchi, F. & Geuking, M. B. Detrimental effect of systemic antimicrobial CD4(+) T-cell reactivity on gut epithelial integrity. Immunology 150, 221–235 (2017).
Solomon, B. D. & Hsieh, C. S. Antigen-specific development of mucosal Foxp3+RORgammat+ T cells from regulatory T cell precursors. J. Immunol. 197, 3512–3519 (2016).
Wegorzewska, M. M. et al. Diet modulates colonic T cell responses by regulating the expression of a Bacteroides thetaiotaomicron antigen. Sci. Immunol. 4, eaau9079 (2019).
Nindl, V. et al. Cooperation of Th1 and Th17 cells determines transition from autoimmune myocarditis to dilated cardiomyopathy. Eur. J. Immunol. 42, 2311–2321 (2012).
Han, B. et al. Developmental control of CD8 T cell-avidity maturation in autoimmune diabetes. J. Clin. Invest 115, 1879–1887 (2005).
Acknowledgements
M.B.G. is supported by a grant (PJT-156073) from the Canadian Health Research Institutes (CIHR) and is a member of the Canadian Microbiome Research Core IMPACTT. We would like to thank Carolyn Thomson, Lukas Mager, and Kathy McCoy for carefully reading the manuscript and their comments.
Author information
Authors and Affiliations
Contributions
M.B.G. and R.B. contributed equally to the writing of this manuscript. R.B. generated the drafts for the figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Geuking, M.B., Burkhard, R. Microbial modulation of intestinal T helper cell responses and implications for disease and therapy. Mucosal Immunol 13, 855–866 (2020). https://doi.org/10.1038/s41385-020-00335-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-020-00335-w
This article is cited by
-
From germ-free to wild: modulating microbiome complexity to understand mucosal immunology
Mucosal Immunology (2022)
-
The impact of the gut microbiota on T cell ontogeny in the thymus
Cellular and Molecular Life Sciences (2022)