Abstract
Mucosal barriers segregate commensal microbes from the intestinal epithelia to maintain gut homeostasis. Ly6/Plaur domain-containing 8 (Lypd8), a highly glycosylated glycosylphosphatidylinositol-anchored protein selectively expressed on colonic enterocytes, promotes this segregation by inhibiting bacterial invasion of the inner mucus layer and colonic epithelia. However, it remains unclear whether Lypd8 prevents infection with enteric bacterial pathogens. Here, we demonstrate that Lypd8 strongly contributes to early-phase defense against Citrobacter rodentium, which causes colitis by inducing attachment and effacement (A/E) lesions on colonic epithelia. Lypd8 inhibits C. rodentium attachment to intestinal epithelial cells by binding to intimin, thereby suppressing the interaction between intimin and translocated intimin receptor. Lypd8 deficiency leads to rapid C. rodentium colonization in the colon, resulting in severe colitis with Th17-cell and neutrophil expansion in the lamina propria. This study identifies a novel function for Lypd8 against A/E bacteria and highlights the role of enterocytes as crucial players in innate immunity for protection against enteric bacterial pathogens.
Similar content being viewed by others
Introduction
A variety of microorganisms gain passage into the mammalian gastrointestinal tract during the intake of food and water. These microorganisms include pathogenic bacteria such as Shigella, Salmonella, and Campylobacter species, along with enteropathogenic and enterohemorrhagic Escherichia coli (EPEC and EHEC, respectively). All of these pathogens cause intestinal inflammation accompanied by severe diarrhea and abdominal pain. Among the enteric pathogens, EHEC and Citrobacter rodentium cause severe colitis following the formation of characteristic pedestal-like structures, termed attaching and effacing (A/E) lesions, on intestinal epithelial cells (IECs). A/E lesions promote tighter binding of A/E bacteria to the epithelia.1 Worldwide, ~3 million people are affected by pathogenic E. coli infection, including EHEC strains, each year.2
To protect the intestinal surface from enteric bacterial pathogens, including A/E bacteria, IECs construct a mucosal barrier mainly composed of mucus layers and antimicrobial peptides.3 These elements have anticolonization and antibacterial effects against pathogenic bacteria, preventing their attachment to the intestinal epithelium or killing the pathogens outright.4 Therefore, mice lacking key components of the mucosal barrier, including mucin and antimicrobial peptides, are highly susceptible to enteric bacterial infection.5,6,7,8 These well-known barrier molecules are mainly produced by goblet cells and Paneth cells, which specialize in the production of mucin and antimicrobial peptides, respectively. It was recently reported that enterocytes produce a unique barrier molecule called Ly6/Plaur domain-containing 8 (Lypd8).9 Lypd8 is a highly N-glycosylated glycosylphosphatidylinositol-anchored protein that is highly expressed on colonic IECs and constitutively shed into the intestinal lumen. Lypd8 inhibits the invasion of colonic epithelia by flagellated commensal bacteria such as Proteus mirabilis and E. coli belonging to Enterobacteriaceae family by suppressing their motility via flagella binding. However, it remains unclear whether Lypd8 has a protective function against pathogenic bacteria, including pathogenic E. coli, Salmonella, Shigella, and Citrobacter, many of which belong to the family Enterobacteriaceae.
A/E bacteria invading the gut mucosa with high motility express type III secretion system (T3SS), which forms a link between the bacterial cytoplasm and the host cell cytoplasm.10 Following maturation of the T3SS translocon, translocated intimin receptor (Tir) is transported and inserted into the host cell membrane, where it interacts with its ligand, intimin.11 The interaction between intimin and Tir allows the intimate attachment of A/E bacteria to IECs, resulting in pedestal formation and actin polymerization.12 With regard to the host immune systems against A/E bacteria, several reports have demonstrated that interleukin (IL)-17 and IL-22 from helper T cells activated by bacterial attachment to IECs play important roles in protecting the mucosa from A/E bacterial infection by stimulating the secretion of antimicrobial molecules by the IECs.13,14,15,16
In this study, we identified a novel innate defense mechanism against A/E bacteria mediated by Lypd8 expressed on enterocytes at the uppermost epithelial layer of the colon, the site first exposed to gut-invading A/E bacteria. Lypd8 repressed C. rodentium attachment to IECs by binding to intimin expressed on the bacterial cell surface. In Lypd8-deficient mice, C. rodentium rapidly colonized the colon and invoked severe colitis, the result of abundant bacterial attachment to IECs. These findings increase our understanding of the defense mechanisms by enterocytes against enteric bacterial infection.
Results
Lypd8-deficient mice are sensitive to C. rodentium infection
Lypd8 contributes to the segregation of commensal bacteria from colonic epithelia in a steady state by suppressing flagellated bacterial invasion. However, the role of Lypd8 in pathogenic bacterial infection is unknown. To determine the role of Lypd8 in protection against enteric pathogens, we first tested the ability of orally infected Lypd8−/− mice to clear C. rodentium. We evaluated the numbers of C. rodentium (measured in colony-forming units, CFU) in fecal samples from Lypd8−/− mice over the course of infection (Fig. 1a). Strikingly, C. rodentium was significantly more abundant in the feces of Lypd8−/− mice compared with wild-type (WT) mice at the early time points, indicating that C. rodentium more rapidly colonized and proliferated in the colons of Lypd8−/− mice. Some probiotic intestinal bacteria prevent the colonization and overgrowth of pathogenic species17; however, a previous work revealed no alterations in the gut microbial community composition of Lypd8−/− mice,9 suggesting that gut bacteria are not involved in the rapid colonization and overgrowth of C. rodentium in Lypd8−/− mice. Accordingly, 4 week-cohousing with WT mice, normalizing gut microbes, did not affect the rapid colonization in Lypd8−/− mice (Supplementary Fig. 1). Intimin-mediated bacterial attachment to IECs is necessary for colonization of A/E bacteria.18 Indeed, intimin-deficient C. rodentium did not colonize in the colons of either WT or Lypd8−/− mice (Supplementary Fig. 2). Thus, we next analyzed the attachment of C. rodentium to colonic epithelia of Lypd8−/− mice by assessing CFU counts of C. rodentium adhered to the colon (vigorously washed colon tissues) (Fig. 1b). The total number of adherent C. rodentium was significantly increased in Lypd8−/− mice compared with WT mice from day 3 to day 12 post-infection (p.i.). Mice were then orally infected with green fluorescent protein (GFP)-expressing C. rodentium. Fluorescence microscopy analysis of the washed colon tissues on day 3 and 9 p.i. revealed very few GFP-expressing bacteria in WT mice, while a number of C. rodentium were observed on the mucosal surface of the colons of Lypd8−/− mice (Fig. 1c).
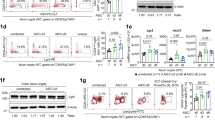
Lypd8 protects colonic mucosa from C. rodentium infection. WT and Lypd8−/− mice were orally infected with 109 CFU of C. rodentium and analyzed on day 12 p.i., unless otherwise stated. a Time course analysis of C. rodentium CFU counts in feces from WT and Lypd8−/− mice. Results are pooled data from three separate experiments, n = 14 mice per group. b Time course analysis of the number (CFU) of C. rodentium cells adhered to colonic tissues (vigorously washed colons) from WT and Lypd8−/− mice. Results are pooled data from two separate experiments, n = 5–10 mice per group. c Fluorescence microscopy imaging of colon sections from WT and Lypd8−/− mice on day 3 and 9 following infection with GFP-expressing C. rodentium. Scale bar, 100 μm. d Transmission electron microscopy images of colon tissues from WT and Lypd8−/− mice. Red arrows indicate A/E lesions underneath adherent bacteria. The area within the yellow dotted line shows bacteria internalized within an epithelial cell. Scale bar, 5 μm. e Hematoxylin and eosin-stained distal colon sections from WT and Lypd8−/− mice show increased thickening of the mucosa and infiltration of cells in Lypd8−/− mice. Scale bar, 100 μm. f Histopathology scores from distal colon tissues of WT and Lypd8−/− mice. Each bar shows scores for damage to the lumen, epithelium, mucosa, and submucosa. Results are pooled data from three separate experiments. Data represent the mean ± SD (n = 15 and 18 per group). A two-tailed Student's t-test was performed to determine statistically-significant differences between WT and Lypd8−/− mice. *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001
Transmission electron microscopy analyses of C. rodentium-infected mice revealed the presence of many A/E lesions beneath the attached bacteria, as well as internalization of bacteria into IECs in the distal colon of Lypd8−/− mice on day 12 p.i. (Fig. 1d). These findings indicate that Lypd8 inhibits the colonization of C. rodentium in the colon by suppressing bacterial attachment to colonic epithelia. In accordance with the increased number of C. rodentium adhering to colonic epithelia, Lypd8−/− mice showed signs of more severe inflammation, characterized by a thicker mucosa, increased cellular infiltration to the lamina propria, and greater submucosal edema (Fig. 1e, f). Taken together, these results suggest that Lypd8 deficiency impairs host defenses against C. rodentium.
To investigate the immune response to C. rodentium infection in Lypd8−/− mice, we first analyzed the expression of reactive oxygen species (ROS)-associated genes such as Nos2, Duoxa2, and Duox2 in colonic epithelial cells, all of which are known to be induced by C. rodentium attachment (Fig. 2a).16 The expression of these genes was higher in the colonic epithelia of uninfected Lypd8−/− mice than in uninfected WT mice. This result is compatible with the fact that a larger number of commensal flagellated bacteria attached to the colonic epithelia of Lypd8−/− mice than was observed in WT mice. Further, the expression of these genes was higher in Lypd8−/− mice than in WT mice following C. rodentium infection. ROS produced by IECs reportedly promote the induction of Th17 cells in the lamina propria following C. rodentium infection.16 Therefore, we next examined the number of helper T cell subsets in the colon tissues of infected mice. The abundance of IL-17+ CD4+ T cells and IFNγ+ IL-17+ CD4+ T cells was dramatically increased in the colons of Lypd8−/− mice compared with WT mice following C. rodentium infection (Fig. 2b and Supplementary Fig. 3A). In addition, the expression of Cxcl1 and Cxcl2 in colonic epithelia was substantially enhanced in Lypd8−/− mice in response to C. rodentium infection, and greater numbers of neutrophils were recruited in C. rodentium-infected Lypd8−/− mice compared with their WT counterparts (Fig. 2c, d and Supplementary Fig. 3B). Thus, Lypd8−/− mice showed enhanced inflammatory responses following C. rodentium infection.
Immune responses to C. rodentium infection in Lypd8−/− mice. WT and Lypd8−/− mice were orally infected with 109 CFU of C. rodentium and analyzed on day 12 p.i. All results are pooled data from two separate experiments. a Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)-based analysis of the expression of ROS-associated genes in colonic epithelial cells from uninfected and C. rodentium-infected WT and Lypd8−/− mice (n = 4 per uninfected group and 10 per infected group). Values were normalized to the expression of Gapdh. b Absolute numbers of CD4+ T cells, Th1 (IFNγ+) cells, Th17 (IL-17+) cells, and INFγ-producing Th17 (IFNγ+ IL-17+) cells in the colon (n = 7 per uninfected group and 6 per infected group). c qRT-PCR assay showing the expression of Cxcl1 and Cxcl2 relative to Gapdh in colonic epithelial cells from C. rodentium-infected and uninfected WT and Lypd8−/− mice (n = 7 per uninfected group and 10 per infected group). d Flow cytometry analysis of the abundance of neutrophils in the colon of uninfected and C. rodentium-infected WT and Lypd8−/− mice (n = 6–8 per group). Data represent the mean ± SD. A two-tailed Student’s t-test was performed for comparison of results between WT and Lypd8−/− mice. *p < 0.05, ***p < 0.005, ****p < 0.001, n.s: not significant
Lypd8 suppresses the attachment of A/E bacteria to colonic epithelia by binding to bacterial cells
We previously demonstrated that Lypd8 hinders the motility of flagellated bacteria including P. mirabilis and E. coli by binding to their flagella. C. rodentium is a member of the family Enterobacteriaceae, most of which are flagellated bacteria. In addition, C. rodentium contains lateral flagellar flagellin gene lafA within its genome, although it does not show obvious motility in vitro culture systems as a result of the deletion of polar flagellar flagellin gene fliC by prophage insertion.19,20 To determine whether the flagellin gene product is targeted by Lypd8 in the defense against C. rodentium invasion of colonic epithelia, we examined the susceptibility of Lypd8−/− mice to infection by lateral flagella-deficient (lafA-deficient, ΔlafA) C. rodentium. When mice were orally infected with an equal number of wild-type and ΔlafA C. rodentium, the proportion of ΔlafA strain in fecal bacteria was less than that of wild-type strain, suggesting that the ΔlafA mutant strain was less competitive and virulent than the wild-type strain (Supplementary Fig. 4A, B). However, the ΔlafA mutant colonized the colons of wild-type mice and induced expansion of Th17 cells and neutrophils at levels comparable with those of wild-type C. rodentium (Supplementary Fig. 5A–C). Unexpectedly, a larger number of ΔlafA cells attached to colonic epithelia of Lypd8−/− mice than wild-type mice, and the ensuing colitis was more severe in Lypd8−/− mice compared with wild-type mice, as was observed following infection with the wild-type strain (Fig. 3a–d). In addition, the expansion of Th17 cells and the recruitment of neutrophils were increased in the colons of Lypd8−/− mice following infection with the ΔlafA C. rodentium strain (Fig. 3e, f). These findings indicate that the inhibitory function of Lypd8 against flagella-mediated bacterial motility is not the main mechanism of resistance to C. rodentium infection.
Lateral flagella deficiency did not affect the susceptibility of Lypd8−/− mice to C. rodentium. WT and Lypd8−/− mice were orally infected with 109 CFU of ΔlafA C. rodentium and analyzed on day 12 p.i., unless otherwise stated. a Number of ΔlafA C. rodentium cells adhering to colonic tissues (vigorously washed colons) from WT and Lypd8−/− mice (n = 5 per group). b Fluorescence microscopy images of GFP-expressing ΔlafA C. rodentium cells in colon sections from WT and Lypd8−/− mice on day 9 p.i. Scale bar, 100 μm. c Hematoxylin and eosin staining of colon sections from ΔlafA C. rodentium-infected WT and Lypd8−/− mice. Scale bar, 100 μm. d Histopathology scores from distal colon tissues of WT and Lypd8−/− mice infected with ΔlafA C. rodentium. e Flow cytometric analysis of CD4+ T cell subsets and neutrophils in the colons of mice infected with ΔlafA C. rodentium. Data represent the absolute numbers of indicated cell types (n = 4 per uninfected group and 9 per infected group). Data represent the mean ± SD. A two-tailed Student’s t-test was conducted for comparison between WT and Lypd8−/− mice. *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001
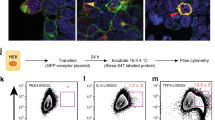
To understand the mechanism by which Lypd8 inhibits the attachment of C. rodentium to the colonic epithelia, we next tested whether Lypd8 binds to C. rodentium in the colon. Wild-type mice were orally infected with C. rodentium. At day 12 p.i., colon tissues were harvested and fixed with Carnoy’s fixative, which preserves luminal contents. The colon sections were then stained with anti-C. rodentium antibody and anti-Lypd8 antibody and analyzed by confocal microscopy (Fig. 4a). C. rodentium was detected on the surface of epithelial cells and at the interface of the inner and outer mucus layer, where Lypd8 colocalized. Therefore, we next used flow cytometry with anti-Lypd8 antibody to analyze Lypd8 binding to GFP-expressing C. rodentium in the feces of WT mice following infection (Fig. 4b). Lypd8 bound to GFP-expressing C. rodentium. C. rodentium attachment is strengthened by the generation of A/E lesions, which are characterized by the destruction of brush border microvilli and membrane protrusions underneath the attached bacteria. At the initiation of A/E lesion formation, intimin (encoded by eae) expressed on the bacterial surface interacts with Tir on the host cell membrane after being injected via the T3SS. Intimin contains a C-terminal C-type lectin domain, which has the capacity to bind carbohydrates of a glycosylated protein.21,22 The T3SS has a similar structure to flagella.23 Because Lypd8 is a highly glycosylated protein that binds to flagella, we generated eae-deficient (Δeae; equivalent to intimin-deficient) and escN-deficient (ΔescN; equivalent to T3SS-deficient) C. rodentium strains and examined Lypd8 binding. Mice were orally infected with the individual mutant strains and subsequently analyzed for bacterial abundance in the feces, the expression of ROS-associated genes, and the abundance of Th17 cells and neutrophils in the colon (Supplementary Fig. 6A–C). Neither of the mutant strains showed increased abundance in the feces of infected mice compared with the WT bacterial strain and, accordingly, they did not induce an increase in expression of ROS-associated genes or an expansion of Th17 cells and neutrophils. To overcome the decreased colonization ability of these strains, we kept on administering orally throughout the infection two antibiotics, vancomycin and chloramphenicol, to which the mutant GFP-expressing strains were resistant (Supplementary Fig. 7), effectively creating a gut environment in which the mutant strains could be competitive. After 2 weeks of treatment with the two antibiotics, we orally infected mice with GFP-expressing wild-type, Δeae, ΔescN, and ΔlafA C. rodentium strains and analyzed the rate of Lypd8 binding to the bacteria in feces on day 12 p.i. by flow cytometry (Fig. 5a–c). Lypd8 binding to the flagella-deficient strain was reduced, implying that Lypd8 binds to lateral flagella of C. rodentium in vivo. In contrast, no decrease in Lypd8 binding to the T3SS-deficient strain compared with the WT was observed, indicating that T3SS is not a target of Lypd8. Intriguingly, Lypd8 binding to the intimin-deficient strain was substantially reduced compared with the WT strain. Lypd8 binding to the intimin and flagella double-mutant C. rodentium strain was severely reduced (Fig. 5d). These findings suggest that intimin on the surface of C. rodentium is targeted by Lypd8 in the colon.
Lypd8 binds to C. rodentium in the colon. a Immunostaining of colon sections fixed in Carnoy’s solution from WT mice on day 12 p.i. using anti-Lypd8 antibody (red) and anti-C. rodentium antibody (green). Scale bars, 20 μm. b Flow cytometric analyses of fecal bacteria from WT and Lypd8−/− mice on day 12 p.i. with GFP-expressing C. rodentium using anti-Lypd8 antibody. The boxed regions indicate the GFP-expressing C. rodentium population in fecal bacteria. The Lypd8-bound GFP-expressing C. rodentium population in fecal bacteria from WT mice is shown in the upper right panel
Lypd8 binding to intimin-deficient C. rodentium was substantially reduced. a In the course of antibiotic (vancomycin and chloramphenicol) treatment, WT and Lypd8−/− mice were orally infected with WT or lafA, escN, or eae-deficient strains expressing GFP. Lypd8 binding to each C. rodentium strain in fecal samples was analyzed by flow cytometry using anti-Lypd8 antibody. b Lypd8 binding to each GFP-expressing bacterial strain in fecal bacteria is shown using histogram charts. c Bars indicate the percentage of Lypd8 binding to each strain (n = 3 per group). Data are shown as mean ± SD. One-way ANOVA was used to determine a significant difference between groups. *p < 0.05, n.s: not significant. d Lypd8 binding to the eae/lafA double-mutant strain is shown using histogram charts
Lypd8 interferes with the interaction between intimin and Tir of A/E bacteria by binding to intimin
To further define Lypd8 binding to intimin expressed by C. rodentium, we used an enzyme-linked immunosorbent assay (ELISA) with anti-FLAG antibody to analyze whether FLAG-tagged Lypd8 recombinant protein could bind to GST-tagged intimin recombinant protein. The Ig-like and C-type lectin (CTL) domains of intimin are pivotal for intimin binding to Tir of A/E bacteria.24 Thus, we examined the binding of Lypd8 to each separate intimin protein domain (Fig. 6a). Predictably, highly glycosylated Lypd8 protein bound to the CTL domain of intimin (Fig. 6b), but also bound to the Ig-like domain. In agreement with our previous study outlining Lypd8 binding to the flagella of E. coli and P. mirabilis,9 Lypd8 did not associate with the C. rodentium flagellin protein encoded by lafA. In addition, human protein LYPD8 bound to intimin from EHEC (Fig. 6c). We further investigated the binding kinetics of Lypd8 and intimin by surface plasmon resonance analysis using the Biacore system (Fig. 6d and Supplementary Fig. 8). A curvature of the binding response in the association phase and a decay in the dissociation phase were observed for the combinations of mouse Lypd8/C. rodentium intimin and human Lypd8/EHEC intimin. To address the mechanism by which Lypd8 binds to intimin, we generated non-glycosylated Lypd8 protein (Supplementary Fig. 9) and analyzed the binding of this protein to each domain of the intimin protein by ELISA (Fig. 6e). As expected, non-glycosylated Lypd8 protein did not associate with the intimin CTL domain, but could still bind to the Ig-like domain. These findings suggest that Lypd8 binds to intimin through both glycan-lectin and protein–protein interactions.
Lypd8 associates with intimin. a Schematic showing the domain structures of the C. rodentium and EHEC intimin proteins. b, c Enzyme-linked immunosorbent assays (ELISA) were performed to analyze the binding of FLAG-tagged Lypd8 and GST-tagged intimin. Increases in the optical density (OD), indicating mouse Lypd8 binding to each of the C. rodentium intimin domains, are shown by a line graph (b). The binding of human LYPD8 to EHEC intimin was also confirmed by increases in the OD (c). d Sensorgram of Biacore system investigating the binding between mouse Lypd8 and C. rodentium intimin and calculated kinetic parameters were shown. e ELISA analysis of the binding between unglycosylated mouse Lypd8 and individual domains of the intimin proteins from C. rodentium. b, c, and e Data represent the mean ± SD of triplicate samples. Representative results from three independent experiments are shown
The interaction between intimin and Tir is indispensable for the generation of A/E lesions.1 To clarify the mechanism by which Lypd8 suppresses the generation of A/E lesions, we used ELISA to examine whether Lypd8 inhibits the interaction between intimin and Tir (Fig. 7a, b). High absorbance readings indicative of intimin-Tir binding gradually decreased following the addition of increasing concentrations of Lypd8. Binding between Tir and the CTL or Ig-like domains of intimin was also negatively impacted by the addition of Lypd8 (Fig. 7c–e). We then further investigated the inhibitory role of mouse Lypd8 on the interaction between intimin and Tir by surface plasmon resonance analysis (Fig. 7f). The value of the association rate constant (Ka) between intimin and Tir was decreased by the addition of Lypd8 protein relative to the control. These results indicate that Lypd8 inhibits the association between intimin and Tir.
Lypd8 inhibits the interaction between intimin and Tir from C. rodentium. a, b The effect of Lypd8 on the intimin-Tir interaction was assessed by ELISA. Changes in the OD, indicative of the binding of intimin to Tir in the presence of increasing concentrations of Lypd8, are shown in the line graph (a) and the bar graph (b). c–e The inhibitory function of Lypd8 on the interactions between the C-terminal domains of intimin (C-type lection domain (CTL) and Ig-like domain (Ig)) and Tir was analyzed by ELISA. The decrease in OD following the addition of increasing concentration of Lypd8 is shown in the line graph (c) and the bar graphs (d and e). Data represent the mean ± SD of triplicate samples. Representative results from three independent experiments are shown. f Sensorgrams of Biacore system investigating the binding of C. rodentium intimin and Tir in the presence of mouse Lypd8 protein and calculated kinetic parameters were shown. A two-tailed Student’s t-test was performed for comparison between groups. *p < 0.05, ***p < 0.005, ****p < 0.001
Discussion
This study reveals a novel immune function for enterocyte-derived Lypd8 against enteric bacterial pathogens. Lypd8 selectively expressed on enterocytes of the colon directly associated with invasive C. rodentium in the region just above colonic epithelia. Lypd8 suppresses the attachment of C. rodentium to IECs by inhibiting the interaction between intimin and Tir by competitively binding to intimin, effectively blocking the interaction that is indispensable for the generation of A/E lesions. In mice lacking Lypd8, a larger number of C. rodentium attached to the colonic epithelia and rapidly colonized the colon, leading to more severe Th17-mediated and neutrophil-mediated colitis accompanied by increased ROS production by colonic epithelial cells.
Mucosal barriers, which mainly comprise mucus layers and antimicrobial peptides generated by goblet cells and Paneth cells, respectively, suppress the colonization and overgrowth of pathogenic bacteria.5,25 We previously demonstrated that Lypd8 expressed by colonic enterocytes inhibits the invasion of commensal flagellated bacteria such as P. mirabilis by suppressing their motility via flagella binding.9 Flagella-mediated motility plays an important role in the initial stages of infection of many bacterial pathogens.26 This implies that Lypd8 has the potential to inhibit the colonization and invasion of colon tissues by enteric bacterial pathogens such as C. rodentium and pathogenic E. coli. Indeed, C. rodentium more rapidly colonized the colon and induced more severe colitis in Lypd8−/− mice. Lypd8−/− mice showed a sign of mild colonic inflammation at the steady state, which might affect the susceptibility to C. rodentium infection. In particular, the damage of the inner mucus layer in Lypd8−/− mice may be involved in the increasing susceptibility to C. rodentium. However, Lypd8 directly bound to intimin of C. rodentium, suggesting that Lypd8 has a direct inhibitory function on the generation of A/E lesion by C. rodentium. On the other hand, C. rodentium was equally eliminated by Lypd8−/− and WT mice in the late stage of infection. This might be the result of a higher number of Th17 cells and neutrophils in the lamina propria at this time point, which contributes to the elimination of C. rodentium.27,28
In vitro-cultured C. rodentium do not express visible polar flagella and, accordingly, are non-motile in soft agar medium. This is most likely because flagellin gene fliC is disrupted by prophage CRP20 in the C. rodentium genome.19,20 However, lateral flagella-associated genes, including lateral flagellar flagellin gene lafA, remain intact. Thus, we examined the effects of Lypd8 on lateral flagella-deficient C. rodentium in the colon. Binding of Lypd8 to the lateral flagella-deficient strain was reduced compared with binding to the WT strain, implying that C. rodentium expresses lateral flagella in the colon, and that they are targeted by Lypd8. A previous report showed that the flagella of EPEC strains are directly involved in adherence to epithelial cells.29 Moreover, another study demonstrated that a lateral flagella-deficient Aeromonas caviae strain had a decreased ability to adhere to Caco-2 cells.30 Nevertheless, C. rodentium lacking lateral flagella could still adhere to colonic epithelia and induce intestinal inflammation, and the number of adherent bacteria in Lypd8−/− mice was higher than that in WT mice following infection with the ΔlafA C. rodentium strain. Although several in vitro analyses using epithelial cell lines have demonstrated the important role of flagella in bacterial attachment, our in vivo analyses suggest that lateral flagella are redundant in the attachment and colonization of C. rodentium in the colon, and that there is another Lypd8-target molecule on the surface of C. rodentium.
Intimin and the T3SS expressed on the surface of C. rodentium are essential players in the generation of A/E lesions. These two components were candidate targets for Lypd8 because the structure of the T3SS is similar to that of the flagella,23 while intimin contains a C-type lectin domain in the C-terminal region that is predicted to bind to a variety of carbohydrate ligands.21,22 We determined that mouse Lypd8 binds to intimin from C. rodentium but does not bind to the T3SS. Furthermore, we found that human Lypd8 also binds to EHEC intimin, indicating that the ability of Lypd8 proteins to associate with intimin from A/E bacteria is conserved in humans. Interestingly, the binding of Lypd8 to intimin is mediated not only by glycan-lectin interactions, but also by protein-protein interactions. Previous studies on the crystal structure of EHEC intimin demonstrated that both the Ig-like and CTL domains of intimin play a role in binding to Tir, with four residues (S890, T909, N916, and N927) of the CTL domain shown to be essential for Tir binding.31,32 This suggests the possibility that Lypd8 bridges the four residue-enclosed pocket essential for Tir binding through its association with the two intimin domains. To better understand the mechanism of the inhibitory function of Lypd8 in disrupting the intimin-Tir association, molecular structure analyses of Lypd8 should be conducted.
This study reveals the role of Lypd8 expressed by colonic enterocytes in providing early-phase protection against enteric bacterial pathogens. Enterocytes, previously regarded as passive contributors to the defense against invading pathogens compared with other intestinal epithelial cells such as goblet cells, Paneth cells, and tuft cells, actively contribute to the fight against enteric pathogens by expressing Lypd8. Further structural analyses to determine the critical protein domain or carbohydrate structure of Lypd8 for binding to intimin should be conducted, and may aid in the development of a novel non-antibiotic therapy for the treatment of EPEC and EHEC infections, which often have severe complications such as hemolytic uremic syndrome.
Materials and methods
Mice
Lypd8−/− mice have been described previously.9 C57BL/6J mice were purchased from Japan SLC (Shizuoka). Lypd8−/− mice were backcrossed to C57BL/6J mice for at least 10 generations. Eight-week-old to twelve-week-old wild-type and Lypd8−/− littermates were used for all in-vivo experiments. All mice were housed under SPF conditions at the Experimental Animal Facility, Graduate School of Medicine, Osaka University. All animal experiments were performed according to the guidelines of the Animal Research Committee of the Graduate School of Medicine at Osaka University.
C. rodentium infection of mice
Mice were infected by oral gavage with 0.4 mL of an overnight culture of Luria-Bertani (LB) broth containing ~1 × 109 colony-forming units (CFU) of C. rodentium NBRC 105723T. Feces were collected on days 3, 6, 9, 12, and 16 post-infection (p.i.). Colons were isolated on day 12 p.i. and vigorously washed in three times with phosphate-buffered saline (PBS). Collected feces and colon tissues were weighed and homogenized. Serial dilutions of the homogenates were prepared in PBS and spread-plated onto MacConkey agar (Merck). Following incubation at 37 °C for 16 h, colonies of the appropriate dilutions were counted, and CFU/g of feces or colonic tissue were calculated. C. rodentium was identified by the generation of small, pink colonies. Vigorously washed colons from mice infected with GFP-expressing C. rodentium were fixed in 4% paraformaldehyde. Frozen sections of the fixed colons were stained and mounted using Fluoro-KEEPER Antifade Reagent with 4ʹ,6-diamidino-2-phenylindole (DAPI; Nacalai Tesque). Carnoy’s solution-fixed sections were stained with rabbit anti-Citrobacter koseri antibody (cat. #ab37056, Abcam)/Alexa Fluor 594 goat anti-rabbit IgG (cat. #A11012, Invitrogen) and anti-mouse Lypd8 antibody (cat. #SA025A4, Biolegend)/Alexa Fluor 647 goat anti-mouse IgG1 (cat. #A21240, Invitrogen), before being counterstained with 4ʹ,6-diamidino-2-phenylindole (DAPI; Vector Laboratories). The stained sections were analyzed using a confocal microscope (FV1000-D; Olympus).
Transmission electron microscopy
Unwashed distal colons from mice infected with C. rodentium were fixed with 2.5% (v/v) glutaraldehyde in 0.1 M phosphate buffer at 4 °C overnight. The colons were post-fixed with 1% (v/v) OsO4 in the same buffer at 4 °C for 1 h, dehydrated using a graded series of ethanol, and embedded in Quetol 812 (Nissin EM). Silver sections were cut using an ultramicrotome, stained with lead citrate and uranyl acetate, and observed under an H-7650 electron microscope (Hitachi).
Histological analysis
Unwashed colons from 8–12-week-old mice were fixed in methanol-Carnoy’s fixative composed of 60% (v/v) methanol, 30% (v/v) chloroform, and 10% (v/v) acetic acid. Paraffin-embedded sections were dewaxed and hydrated. Sections were then stained with hematoxylin and eosin. Images were obtained using a BZ-9000 fluorescence microscope (Keyence). Tissue sections were evaluated for pathology in four regions: lumen, surface epithelium, mucosa, and submucosa. Pathology in the lumen was based on the presence of necrotic epithelial cells (0: none, 1: scant, 2: moderate, 3: dense). The surface epithelium was scored for regenerative change (0: none, 1: mild, 2: moderate, 3: severe), desquamation (0: no change, 1: <10 epithelial cells shedding per lesion, 2: 11–20 epithelial cells shedding per lesion), and ulceration (3: epithelial ulceration, 4: epithelial ulceration with severe crypt destruction). The mucosa was scored for hyperplasia based on crypt length (0: < 200 µm, 1: 200–280 µm, 2: 280–350 µm, 3: > 350 µm) and goblet cell depletion (0: no change, 1: mild, 2: moderate, 3: severe). Last, the submucosa was scored for edema (0: no change, 1: mild, 2: moderate, 3: severe).
Flow cytometric analysis of C. rodentium in fecal bacteria
For the purpose of colonization with avirulent mutant strains, antibiotics were administered to mice by the addition of vancomycin (0.5 g/L; Duchefa Biochemie B.V.) and chloramphenicol (0.5 g/L; Nacalai Tesque) to autoclaved drinking water. Two weeks after the start of antibiotic treatment, mice were orally infected with each GFP-expressing C. rodentium strain. The two antibiotics were orally administered continuously throughout the infection to keep the reduction of commensal bacteria. In the case of assay for competition of wild-type and ΔlafA strains, WT mice were orally infected with the mixture of the wild-type strain expressing GFP (wt (GFP)) and the ΔlafA strain expressing mCherry (ΔlafA (mCherry)). Feces were collected in tubes from mice on day 12 p.i., weighed, and diluted 10-fold in PBS. Diluted specimens were mixed well and centrifuged at 400×g for 5 min to remove bacteria from the larger particles. Aliquots (200 μl) of the supernatant were centrifuged at 8000×g for 10 min, and bacterial pellets were fixed in PBS containing 4% (v/v) paraformaldehyde for an hour. After the removal of fixative, for the assay for Lypd8 binding to C. rodentium, fecal bacteria from both the WT and Lypd8−/− mice were incubated with anti-Lypd8 antibody. After washing with PBS containing 2% (v/v) FBS, bacterial pellets were stained with Alexa Fluor 647 goat anti-mouse IgG1 (Invitrogen). Lypd8 binding to GFP-positive C. rodenitum in bacterial suspensions was analyzed using the FACS Canto II system (BD Biosciences) with FlowJo software (Tree Star). The proportion of wt (GFP) and ΔlafA (mCherry) in fecal bacteria was analyzed by FACS AriaIIIu (BD Biosciences) with FlowJo software (Tree Star).
ELISA
To evaluate Lypd8 binding to bacterial proteins, 96-well plates (Corning) were coated with 10 μg/mL GST-tagged bacterial proteins for 16 h at 4 °C. Plates were blocked with 3% w/v bovine serum albumin (BSA) in PBS, and then incubated with increasing concentrations of FLAG-tagged Lypd8 protein diluted in PBS for 2 h at room temperature. Plates were then washed, and 0.5 μg/mL horseradish peroxidase-conjugated anti-FLAG M2 mAb (Sigma-Aldrich) diluted in PBS was added to each well. Plates were then incubated for 2 h at room temperature before being washed, followed by the addition of TMB substrate. The optical density was then read at 450 nm using a spectrometer.
To evaluate the inhibitory function of Lypd8 against the interaction between intimin and Tir, 96-well plates (Corning) were coated with 10 μg/mL untagged Tir protein for 16 h at 4 °C. Plates were blocked with 3% (w/v) BSA in PBS and then incubated with 10 μg/mL GST-tagged intimin protein mixed with increasing concentrations (0–500 μg/mL) of FLAG-tagged Lypd8 protein for 2 h at room temperature. Plates were then washed, and 0.5 μg/mL horseradish peroxidase-conjugated anti-GST pAb (MBL) diluted in PBS was added to each well, followed by incubation at room temperature for 2 h. Following washing, TMB substrate was added to each well. Optical densities were then read at 450 nm using a spectrometer.
Surface plasmon resonance (SPR) interaction analysis (Biacore)
To assess the binding of Lypd8 and intimin, GST-tagged C. rodentium intimin (Ig-like domain and CTL domain) and GST-tagged EHEC intimin (Ig-like domain and CTL domain) were diluted in 10 mM sodium acetate (pH 5 and 4.5, respectively) and 2000 response units were immobilized via amine coupling to Series S Sensor Chip CM5 flow chambers (GE Healthcare) using an Amine Coupling Kit (GE Healthcare). A flow chamber without protein was used as a control (blank) for each experiment. Recombinant Lypd8 protein was sequentially-diluted in running buffer (10 mM HEPES, 0.15 M NaCl, 0.0025% (v/v) Surfactant P20, pH 7.5) and typically injected over the different surfaces at 10 μL/min.
To assess the interaction between intimin and Tir of C. rodentium in the presence of Lypd8, untagged Tir was diluted in 10 mM sodium acetate (pH 4.5). A total of 1500 response units were immobilized via amine coupling to Series S Sensor Chip CM5 flow chambers using an Amine Coupling Kit (GE Healthcare). A flow chamber without protein was used as a control (blank) for each experiment. Recombinant intimin protein (Ig-like domain and CTL domain) was sequentially diluted in running buffer (10 mM HEPES, 0.15 M NaCl, 0.0025% (v/v) Surfactant P20, pH 7.5) with/without Lypd8 protein and typically injected over the different surfaces at 30 μL/min.
Binding was monitored in a Biacore T200 instrument (GE Healthcare). Between experiments, the surfaces were strictly regenerated with 10 mM glycine-HCl, pH 2.5, followed by an extensive wash procedure using running buffer (10 mM HEPES, 0.15 M NaCl, 0.0025% (v/v) Surfactant P20, pH 7.5). The obtained sensorgram data were evaluated using Biacore T200 evaluation software. The blank bulk refraction curves from the control flow chamber for each injected concentration were subtracted from the results. The blank subtracted sensorgrams were fitted to the 1:1 binding model, and the association (Ka) and dissociation (Kd) rate constants and dissociation rate (KD) were determined.
Statistical analysis
Differences between groups were evaluated by Student's t-test for single comparisons or one-way ANOVA for multiple comparisons using GraphPad Prism software, followed by post-hoc Bonferroni tests. A p-value < 0.05 was considered significant. Data are presented as mean ± SD.
References
Collins, J. W. et al. Citrobacter rodentium: infection, inflammation and the microbiota. Nat. Rev. Microbiol. 12, 612–623 (2014).
Majowicz, S. E. et al. Global incidence of human Shiga toxin-producing Escherichia coli infections and deaths: a systematic review and knowledge synthesis. Foodborne Pathog. Dis. 11, 447–455 (2014).
Maynard, C. L., Elson, C. O., Hatton, R. D. & Weaver, C. T. Reciprocal interactions of the intestinal microbiota and immune system. Nature 489, 231–241 (2012).
Okumura, R. & Takeda, K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp. Mol. Med. 49, e338 (2017).
Bergstrom, K. S. et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 6, e1000902 (2010).
Goto, Y. et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 345, 1254009 (2014).
van Ampting, M. T. et al. Intestinally secreted C-type lectin Reg3b attenuates salmonellosis but not listeriosis in mice. Infect. Immun. 80, 1115–1120 (2012).
Wilson, C. L. et al. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 286, 113–117 (1999).
Okumura, R. et al. Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature 532, 117–121 (2016).
Gaytan, M. O., Martinez-Santos, V. I., Soto, E. & Gonzalez-Pedrajo, B. Type three secretion system in attaching and effacing pathogens. Front. Cell. Infect. Microbiol. 6, 129 (2016).
Kenny, B. et al. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91, 511–520 (1997).
Liu, H., Magoun, L., Luperchio, S., Schauer, D. B. & Leong, J. M. The Tir-binding region of enterohaemorrhagic Escherichia coli intimin is sufficient to trigger actin condensation after bacterial-induced host cell signalling. Mol. Microbiol. 34, 67–81 (1999).
Zheng, Y. et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 (2008).
Backert, I. et al. STAT3 activation in Th17 and Th22 cells controls IL-22-mediated epithelial host defense during infectious colitis. J. Immunol. 193, 3779–3791 (2014).
Aychek, T. et al. IL-23-mediated mononuclear phagocyte crosstalk protects mice from Citrobacter rodentium-induced colon immunopathology. Nat. Commun. 6, 6525 (2015).
Atarashi, K. et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380 (2015).
Sassone-Corsi, M. & Raffatellu, M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J. Immunol. 194, 4081–4087 (2015).
McKee, M. L., Melton-Celsa, A. R., Moxley, R. A., Francis, D. H. & O'Brien, A. D. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect. Immun. 63, 3739–3744 (1995).
Petty, N. K. et al. Citrobacter rodentium is an unstable pathogen showing evidence of significant genomic flux. PLoS Pathog. 7, e1002018 (2011).
Khan, M. A. et al. Flagellin-dependent and -independent inflammatory responses following infection by enteropathogenic Escherichia coli and Citrobacter rodentium. Infect. Immun. 76, 1410–1422 (2008).
Zhang, X. W. et al. A C-type lectin with an immunoglobulin-like domain promotes phagocytosis of hemocytes in crayfish Procambarus clarkii. Sci. Rep. 6, 29924 (2016).
Kelly, G. et al. Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli. Nat. Struct. Biol. 6, 313–318 (1999).
Notti, R. Q. & Stebbins, C. E. The structure and function of type III secretion systems. Microbiol. Spectr. 4, https://doi.org/10.1128/microbiolspec.VMBF-0004-2015 (2016).
Batchelor, M. et al. Structural basis for recognition of the translocated intimin receptor (Tir) by intimin from enteropathogenic Escherichia coli. EMBO J. 19, 2452–2464 (2000).
Loonen, L. M. et al. REG3gamma-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 7, 939–947 (2014).
Mellies, J. L. & Lorenzen, E. Enterohemorrhagic Escherichia coli virulence gene. Regul. Microbiol. Spectr. 2, EHEC-0004–EHEC-2013 (2014).
Mangan, P. R. et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature 441, 231–234 (2006).
Lebeis, S. L., Bommarius, B., Parkos, C. A., Sherman, M. A. & Kalman, D. TLR signaling mediated by MyD88 is required for a protective innate immune response by neutrophils to Citrobacter rodentium. J. Immunol. 179, 566–577 (2007).
Giron, J. A., Torres, A. G., Freer, E. & Kaper, J. B. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44, 361–379 (2002).
Gavin, R. et al. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 43, 383–397 (2002).
Liu, H. et al. Point mutants of EHEC intimin that diminish Tir recognition and actin pedestal formation highlight a putative Tir binding pocket. Mol. Microbiol. 45, 1557–1573 (2002).
Yi, Y. et al. Crystal structure of EHEC intimin: insights into the complementarity between EPEC and EHEC. PLoS One 5, e15285 (2010).
Acknowledgements
We thank T. Kondo and Y. Magota for technical assistance, Y. Fujioka, T. Nakano, and K. Sano at Osaka Medical College for analysis of bacterial morphology, T. Sheen at Edanz Group for editing a draft of this paper, and C. Hidaka for secretarial assistance. The transmission electron microscopic study was supported by E. Oiki and N. Hayakawa at the Center for Medical Research and Education, Graduate School of Medicine, Osaka University. The surface plasmon resonance interaction analysis using Biacore was supported by Y. Ito at the Center for Medical Research and Education, Graduate School of Medicine, Osaka University. This study was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (A18H040280 and T18K151870), the Japan Agency for Medical Research and Development (JP18gm1010004), and the Terumo Foundation for Life Sciences and Arts.
Author information
Authors and Affiliations
Contributions
R.O. planed and performed experiments and wrote the paper. T.K. and T.I. generated C. rodentium mutant strains. C.C.H. and B.H.S. generated recombinant proteins and performed animal experiments. S.H. performed gene expression analyses. T.K. performed animal experiments. K.T. planned and directed the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Okumura, R., Kodama, T., Hsu, CC. et al. Lypd8 inhibits attachment of pathogenic bacteria to colonic epithelia. Mucosal Immunol 13, 75–85 (2020). https://doi.org/10.1038/s41385-019-0219-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-019-0219-4
This article is cited by
-
Intestinal mucus components and secretion mechanisms: what we do and do not know
Experimental & Molecular Medicine (2023)
-
Single-cell transcriptomics reveals intestinal cell heterogeneity and identifies Ep300 as a potential therapeutic target in mice with acute liver failure
Cell Discovery (2023)
-
Gut microbiota alternation under the intestinal epithelium-specific knockout of mouse Piga gene
Scientific Reports (2022)
-
Slimy partners: the mucus barrier and gut microbiome in ulcerative colitis
Experimental & Molecular Medicine (2021)