Abstract
T helper 9 (TH9) cells are important for the development of inflammatory and allergic diseases. The TH9 transcriptional network converges signals from cytokines and antigen presentation but is incompletely understood. Here, we identified TL1A, a member of the TNF superfamily, as a strong inducer of mouse and human TH9 differentiation. Mechanistically, TL1A induced the expression of the transcription factors BATF and BATF3 and facilitated their binding to the Il9 promoter leading to enhanced secretion of IL-9. BATF- and BATF3-deficiencies impaired IL-9 secretion under TH9 and TH9-TL1A-polarizing conditions. In vivo, using a T-cell transfer model, we demonstrated that TL1A promoted IL-9-dependent, TH9 cell-induced intestinal and lung inflammation. Neutralizing IL-9 antibodies attenuated TL1A-driven mucosal inflammation. Batf3−/− TH9-TL1A cells induced reduced inflammation and cytokine expression in vivo compared to WT cells. Our results demonstrate that TL1A promotes TH9 cell differentiation and function and define a role for BATF3 in T-cell-driven mucosal inflammation.
Similar content being viewed by others
Introduction
Specialized subsets of T helper (TH) cells play an integral role in maintaining tissue homeostasis and during the development of inflammatory diseases at mucosal surfaces. TH9 cells have been recently identified as an independent TH cell subset that produces mainly IL-9 but also IL-10 and IL-21.1,2 TH9 cells have been implicated in several diseases, including allergic lung inflammation, experimental autoimmune encephalomyelitis (EAE), colitis, parasitic worm infections, and cancer.1,3,4,5 Recently, TH9 cells have been reported to play a role in inflammatory bowel diseases (IBD).6,7,8,9 Ulcerative colitis (UC) patients have elevated numbers of mucosal IL-9+ T cells and the IL-9 receptor (IL-9R) is upregulated on the intestinal epithelium. IL-9 deficiency suppresses the development of acute and chronic oxazolone-induced colitis, a model that mimics UC.6 In Crohn’s disease (CD) patients, high-serum IL-9 levels correlate with severe disease.8,9,10
TH9 cells differentiate from naive CD4+ T cells in the presence of TGF-β1 and IL-4. Several transcription factors downstream of T-cell receptor (TCR), TGF-β1, and IL-4 receptors are required for the differentiation of TH9 cells including IRF4, STAT6, GATA3, PU.1, NF-κB, and basic leucine zipper transcription factor ATF-like (BATF).1,11,12,13,14,15 However, the transcriptional program and inflammatory triggers that drive the differentiation of TH9 cells are still not well understood and a lineage-defining factor associated with IL-9 expression has not been identified.16
The family of BATF transcription factors is comprised of BATF, BATF2, and BATF3 and belong to the AP-1 family of transcription factors that include JUN and FOS. Transcription factors in the BATF family are composed of a DNA-binding domain and leucine zipper motif but lack a transactivation domain and were initially described as negative regulators of AP-1 activity.17 However, recent studies have demonstrated that BATF family members interact with IRF transcription factors including IRF4 and IRF8, and bind to AP-1-IRF composite element sequences to regulate their target genes in T cells and dendritic cells.18 BATF has been shown to be required for the development of TH9 cells as well as TH2, TH17, and TFH cells.15,19 BATF3 has been described to control the development of CD8α+ and CD103+ dendritic cells.20,21 In contrast to BATF, a nonredundant function of BATF3 in the development of TH cells has not been demonstrated. In addition, the extracellular stimuli that can activate the BATF3 pathway in CD4+ TH cells remain to be elucidated.
We have recently shown that TL1A, a TNF superfamily member that plays an important role in immune-mediated diseases including IBD,22,23,24,25 induces the secretion of IL-9 in TH17 cells.26 Furthermore, a recent publication demonstrated that TL1A, via its receptor DR3, promotes TH9 differentiation through IL-2- and STAT5-dependent but PU.1- and STAT6-independent mechanisms in allergic lung inflammation.27 However, the transcriptional programs induced by TL1A and the pathogenicity of TL1A-induced TH9 cells in intestinal inflammation and IBD remains to be elucidated.
Here, we demonstrate that TL1A promotes the differentiation of human and murine TH9 cells via a novel signaling pathway. We define BATF3 as a novel transcriptional regulator for the differentiation of TH9 cells. TL1A leads to chromatin remodeling at the Il9 locus and recruitment of the pioneer transcription factors IRF4, and BATF, which are important components for IL-9 transcriptional activation, and BATF3 to conserved regions within the Il9 promoter. Furthermore, TL1A upregulates the expression of BATF and BATF3 in a STAT6-dependent manner. Batf- and Batf3-deficient TH9 and TL1A-induced TH9 (TH9-TL1A) cells are impaired in their IL-9 production. TH9-TL1A cells are highly pro-inflammatory in vivo when adoptively transferred into Rag1−/− mice as evident by severe mucosal inflammation in the intestines and lungs, which was attenuated by treatment with neutralizing IL-9 antibodies. Adoptive transfer of Batf3−/− TH9-TL1A cells into Rag1−/− mice leads to significantly reduced mucosal inflammation and cytokine production in comparison with WT TH9-TL1A cells. Our data demonstrate a novel role for TL1A and BATF3 in developing pathogenic effector TH9 cells and identify this signaling pathway as a potential therapeutic target in TH9-driven pathologies, including IBD and allergic lung disease.
Results
TL1A enhances murine and human TH9 cell differentiation in vitro
To determine the effect of TL1A on TH9 differentiation, we stimulated naive CD4+ T cells under TH9 conditions with or without TL1A. TL1A significantly enhanced IL-9 secretion and Il9 mRNA expression (Fig. 1a, Supplementary Figure 1a). TL1A alone did not induce IL-9 secretion, suggesting that TL1A acts synergistically with TGF-β1 and IL-4 to promote IL-9 production. Secretion and mRNA expression of TH9-associated cytokines IL-10 and IL-13 was also enhanced by TL1A (Fig. 1a, Supplementary Figure 1a, b). Consistent with previous reports, time-course experiments of intracellular IL-9 staining revealed transient IL-9 expression with a maximal induction under TH9 conditions at day 3 and a decline back to baseline at day 5 (Supplementary Figure 1b).28,29 TL1A induced a significantly higher percentage of IL-9-producing cells throughout the time course of the experiments with a shift in kinetics toward an earlier and stronger IL-9 induction that even at an early time point exceeded the maximum of IL-9 production induced under TH9 conditions (Supplementary Figure 1b). We also observed an increase of IL-9+IL-10+ cells in the presence of TL1A (Supplementary Figure 1b). We did not observe any shift toward other TH subsets (Supplementary Figure 1c). The enhancement of TH9 differentiation by TL1A was completely dependent on its receptor DR3 (Supplementary Figure 1d). Taken together, these data suggest that TL1A synergizes with TGF-β1 and IL-4 in TH9 differentiation and IL-9 production.
TL1A enhances the differentiation of murine and human TH9 cells. a Naive murine CD4+ T cells were differentiated under TH0- or TH9-polarizing conditions with or without TL1A for 3 days. a ELISA analysis of IL-9, IL-10, and IL-13 secretion. b Naive human CD4+ T cells (CD4+CD45RA+CD45RO−CD25−) were isolated from PBMCs of healthy donors and differentiated under TH9-polarizing conditions with or without TL1A for 3 days. ELISA analysis of human IL-9 secretion at 48 and 72 h. Box-whisker plot showing the IL-9 concentrations in culture supernatants for TH9 and TH9-TL1A-treated cells. The boxes represent the median and 25th and 75th percentile, and the whiskers represent the minimum and maximum values of the distribution. N = 7–9. Data represent means ± SD of one independent experiment out of four (a) independent experiments. *p < 0.05, ***p < 0.005 determined by Student’s t test
TL1A also significantly enhanced TH9 differentiation in an antigen-specific setting using OVA-specific OT-II cells (Supplementary Figure 1e, f). TL1A did not affect cell proliferation during TH9 differentiation (Supplementary Figure 1g), as has been demonstrated for memory CD4+ T cells.30 These data suggest that TL1A enhanced TH9 differentiation without affecting proliferation. Furthermore, we examined IL-9 production in cells that were differentiated under TH9 and TH9-TL1A conditions for 3 days and restimulated with anti-CD3ε/CD28. We observed increased IL-9 production in TH9-TL1A cells compared to TH9 cells after secondary stimulation that was further enhanced in the presence of TL1A (Supplementary Figure 1h, i). These data suggest that TL1A also enhanced IL-9 production from differentiated TH9 cells.
Next, we determined the effect of TL1A on the differentiation of human TH9 cells. Naive CD4+ T cells were isolated from PBMC from healthy donors and stimulated under TH9 condition in the presence of human TL1A. Consistent with our data from mouse TH9 cells, addition of TL1A significantly enhanced IL-9 secretion from human TH9 cell cultures (Fig. 1b).
TL1A induces the expression of the transcription factors BATF and BATF3
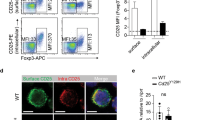
To elucidate signaling pathways induced by TL1A, we used RNA sequencing to assess the transcriptomic profile in TH9 and TH9-TL1A cells. TL1A did not substantially alter the TH9 transcriptional profile indicating that TL1A did not globally skew TH9 differentiation (data not shown). However, 219 genes were differentially expressed in TH9-TL1A cells (Fig. 2a, Supplementary Tables 1 and 2). Pathway enrichment analysis identified significant changes in genes involved in cytokine–cytokine receptor and JAK-STAT signaling pathways in TH9-TL1A conditions (Supplementary Tables 1 and 2, data not shown). BATF3 and several BATF-regulated genes were significantly upregulated under TH9-TL1A conditions (Supplementary Table 3) suggesting that the transcription factors BATF3 and BATF might be involved in the differentiation of TH9-TL1A cells. BATF has been described to be required for the development of TH17 and TH9 cells.15,31,32,33,34 To determine whether TL1A activates BATF family members, we first assessed the expression level of Batf in different TH subsets and detected strong Batf mRNA expression in TH9 and TH2 cells (Fig. 2b). TL1A further enhanced the expression of Batf under TH9 conditions, particularly on day 1 when TH9 conditions alone are not sufficient to induce Batf mRNA (Fig. 2c). TL1A alone induced Batf mRNA to a similar degree as TH9 conditions (Fig. 2c). Intracellular staining for BATF confirmed that TH9-TL1A stimulation significantly increased the percentage of BATF+ cells (Fig. 2d). Furthermore, TL1A alone increased the percentage of BATF+ cells and more importantly the percentage of BATF+IRF4+ cells compared to TH9 conditions suggesting that TL1A might enhance the formation of IRF4-BATF cooperative complexes (Fig. 2d). TH9-TL1A stimulation also enhanced the percentage of BATF+IL-9+ cells (Fig. 2e).
TL1A upregulates BATF and BATF3 expression during TH9 differentiation. a Transcriptional profiling of TH9 and TH9-TL1A cells by RNA sequencing. Heat map displaying RNA-sequencing data using the top 100 genes with the largest IQR (interquartile range). The dendrograms to the left and above the heat map represent clustering of genes (rows) and samples (columns). b Naive CD4+ T cells were differentiated into different TH subsets and Tregs for 3 days. Relative Batf mRNA expression was analyzed by qPCR. c Naive CD4+ T cells were differentiated under TH0- or TH9-polarizing conditions with or without TL1A for the indicated time periods. QPCR analysis of relative Batf mRNA expression. d Co-staining of BATF and IRF4 (top). Quantitative analysis of the percentages of BATF+IRF4+ cells (bottom). e Co-staining of BATF and IL-9. f–g Naive human CD4+ T cells were isolated from PBMCs of healthy volunteers and differentiated under TH9-polarizing conditions with or without TL1A. f Representative intracellular staining of IL-9 and BATF at 48 h. g Frequency of BATF+IL-9+ T cells differentiated under TH9 and TH9-TL1A conditions. Means are shown. Each symbol represents an individual donor. N = 4. h Relative Batf3 mRNA expression in different murine TH subsets and Tregs was analyzed by qPCR. i Relative Batf3 mRNA expression at indicated time points. j Representative co-staining of BATF3 and IL-9. k Frequency of BATF3+IL-9+ T cells differentiated under TH9 and TH9-TL1A conditions. Means are shown. Each symbol represents an individual experiment. N = 3. l–m Naive human CD4+ T cells were differentiated under TH9-polarizing conditions with or without TL1A. l Representative intracellular staining of IL-9 and BATF3 at 48 h. m Frequency of BATF3+IL-9+ T cells differentiated under TH9 and TH9-TL1A conditions. Means are shown. Each symbol represents an individual donor. N = 4. Data represent means ± SD of one independent experiment out of two (a) or at least three (b–d, i, k) independent experiments. *p < 0.05, **p < 0.01, ***p < 0.005 as determined by Student’s t test
In addition, human TH9 cells had significantly higher percentage of IL-9-producing cells when stimulated with TL1A (Fig. 2f, g). Notably, the majority of IL-9-producing cells co-expressed BATF. Co-staining of BATF and IRF4 showed that addition of TL1A increased the percentage of BATF+IRF4+ cells during differentiation of human TH9 (data not shown), suggesting that the enhancement of IL-9 production via the BATF signaling pathway by TL1A is conserved between mouse and human TH9 cells.
In contrast to BATF, the role of BATF3 in the differentiation of TH cells is less defined. To confirm our RNA-seq data, we determined the expression level of Batf3 in TH9 cells and other TH subsets and found that Batf3 mRNA expression was the highest in TH9, TH1, and TH2 cells (Fig. 2h). TL1A enhanced the expression of Batf3 under TH9 conditions (Fig. 2i). Intracellular staining for BATF3 confirmed that TH9-TL1A stimulation significantly increased the percentage of BATF3+ IL-9+ cells (Fig. 2j, k).
In human TH9 cells, TL1A significantly enhanced the expression of BATF3+IL-9+ cells (Fig. 2l, m). Similar to BATF, the majority of IL-9-producing cells co-expressed BATF3.
BATF and BATF3 are critical transcription factors in TL1A-induced TH9 differentiation
To further define the role of BATF and BATF3, we performed chromatin immunoprecipitation (ChIP) assays to determine binding at two Il9 conserved noncoding sequences (Il9 CNS1, CNS0).35 TL1A alone induced significantly greater binding of BATF to Il9 CNS1 compared to TH9 conditions and TH9-TL1A further enhanced BATF binding (Fig. 3a). At the more upstream Il9 CNS0, TL1A also enhanced BATF binding under TH9 conditions while TL1A alone had no effect (Fig. 3b). TL1A enhanced IRF4 binding under TH9 conditions at CNS1 and CNS0 (Fig. 3a, b). Interestingly, binding of acetylated histone 3 (AcH3) at CNS1 and CNS0 that was previously described to facilitate chromatin remodeling at the Il9 locus during TH9 differentiation14 was also enhanced under TH9-TL1A conditions (Fig. 3a, b). BATF3 binding to CNS1 was upregulated in TH9 cells and the binding was further enhanced under TH9-TL1A conditions while BATF3 did not bind to CNS0 (Fig. 3a, data not shown). These data suggest that TL1A enhances BATF, BATF3, and IFR4 binding to the Il9 promoter under TH9 conditions to induce Il9 expression.
TL1A enhances binding of BATF, IRF4, and BATF3 to the Il9 promoter during TH9 differentiation. a, b Naive murine CD4+ T cells were differentiated under TH0- or TH9-polarizing conditions with or without TL1A for 2 (BATF, IRF4, and AcH3), or 3 (BATF3) days. ChIP assays were performed for BATF, IRF4, BATF3, or AcH3 binding to conserved noncoding sequences (CNS) 1 (a) and CNS0 (b) of Il9. Data represent means ± SD of one independent experiment out of two independent experiments. *p < 0.05, **p < 0.01, ***p < 0.005 as determined by Student’s t test
To confirm the roles for BATF and BATF3 in TL1A-dependent TH9 differentiation, we used Batf−/− and Batf3−/− T cells. IL-9, IL-10, and IL-13 production was severely impaired in Batf−/− cells under TH9 conditions (Fig. 4a–d). However, TL1A stimulation at least partially overcame the requirement for BATF for IL-9 and IL-13 production during TH9 differentiation (Fig. 4a, b, d). In the absence of BATF3, the induction of IL-9 was significantly reduced, particularly under TH9-TL1A conditions (Fig. 4e, f, Supplementary Figure 2a). In contrast to BATF, BATF3 was not required for IL-10 or IL-13 expression (Fig. 4g, h, Supplementary Figure 2b, c). To define the signaling pathways that lead to BATF and BATF3 upregulation in the presence of TL1A, we used Stat6−/− and p50−/− cells. p50−/− TH9 cells had reduced percentages of BATF+IRF4+ cells and the increase in the percentages of BATF+IRF4+ cells under TL1A stimulation was almost completely abolished (Supplementary Figure 3a, b). Upregulation of Batf and Batf3 mRNA under TH9 conditions was p50-independent, while the enhancement of Batf and Batf3 expression under TH9-TL1A conditions was completely p50-dependent (Supplementary Figure 3c, d). Stat6−/− TH9 cells had reduced percentages of BATF+IRF4+ cells and their increase under TH9-TL1A stimulation was completely abolished (Supplementary Figure 4a, b). Similarly, upregulation of Batf and Batf3 mRNA was completely dependent on STAT6 under TH9 and TH9-TL1A conditions (Supplementary Figure 4c, d). We observed no significant effects of BATF or BATF3 deficiency on the expression of Batf3 and Batf, respectively, suggesting that there is no compensation between the two transcription factors (Supplementary Figure 5). Together, these data suggest that BATF and BATF3 play important and distinct roles during TH9 differentiation in the presence of TL1A and that increased BATF and BATF3 expression under TL1A stimulation is dependent on STAT6 and the canonical NF-κB pathway.
Effects of BATF and BATF3 deficiencies on TH9 differentiation. a–d Naive CD4+ T cells from Batf−/− or WT mice were differentiated under TH0- or TH9-polarizing conditions with or without TL1A for 3 days. a Intracellular staining of IL-9 and IL-10. b ELISA analysis of IL-9 production. c ELISA analysis of IL-10 production. d ELISA analysis of IL-13 production. e–h Naive CD4+ T cells from Batf3−/− or WT mice were differentiated under TH9-polarizing conditions with or without TL1A for 3 days. e Intracellular staining of IL-9 and IL-10. f ELISA analysis of IL-9 production. g ELISA analysis of IL-10 production. h ELISA analysis of IL-13 production. i Transcriptional profiling of WT and Batf3−/− TH9-TL1A cells by RNA sequencing. Heat map displaying RNA-sequencing data of genes differentially expressed with p < 0.01. The dendrograms to the left and above the heat map represent clustering of genes (rows) and samples (columns). Data represent means ± SD of one independent experiment out of two (i) or three (a–h) independent experiments. *p < 0.05, **p < 0.01, ***p < 0.005 determined by Student’s t test
Next, we performed RNA sequencing on WT and Batf3−/− TH9-TL1A cells. Totally, 139 genes were differentially expressed in WT versus Batf3−/− TH9-TL1A cells (Fig. 4i, Supplementary Table 4). Gene ontology pathway enrichment analysis identified significant changes in genes involved in regulation of transcription, cell proliferation, and intracellular signal transduction (Supplementary Tables 5 and 6). These results reveal a role for BATF3 in the differentiation of TH9-TL1A cells.
TH9-TL1A cells are highly pathogenic in vivo
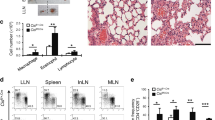
To determine the pathogenic potential of TH9-TL1A cells in vivo, we adoptively transferred ex vivo-differentiated CD45.1+ TH9 or TH9-TL1A cells into Rag1−/− mice. Mice that received TH9 cells did not develop significant weight loss (Fig. 5a). However, mice that received TH9-TL1A cells lost weight and developed more severe intestinal inflammation, particularly in the small intestine (Fig. 5a–c). We also observed severe pulmonary inflammation, characterized by peribronchial and perivascular cellular infiltration of inflammatory cells in TH9-TL1A recipients. Staining with Alcian Blue periodic acid-Schiff (AB-PAS) showed hyperproliferation of mucin-producing cells in the airway epithelium of TH9-TL1A recipients (Supplementary Figure 6a, b). We observed higher cellularity in spleens and lamina propria mononuclear cells (LPMC), and higher percentages and absolute numbers of splenic, MLN, and LPMC CD4+ cells in TH9-TL1A recipients (Fig. 5d, e) suggesting that TH9-TL1A cells have a higher proliferative potential in vivo. Ki67 staining confirmed that cells from MLN, spleen, and LPMC from TH9-TL1A recipients are more proliferative even 6 weeks after T-cell transfers (Fig. 5f). Significantly higher secretion of IL-13 and IL-17 in anti-CD3/anti-CD28-restimulated MLN cells from TH9-TL1A recipients was observed (Fig. 5g), and significantly higher secretion of IL-9 and IL-13 in anti-CD3/anti-CD28-restimulated LPMC from TH9-TL1A recipients (Fig. 5h). We observed a higher absolute number of splenic and MLN IL-9+, IL-13+, and IL-17+ cells and a higher absolute number of MLN, splenic, and LPMC CCR6+ and CD103+ T cells in TH9-TL1A recipients consistent with a role of these cells at mucosal surfaces (Fig. 5i, j).
TH9 cells differentiated in the presence of TL1A induce more severe mucosal inflammation. Naive CD4+ T cells from CD45.1 mice were differentiated under TH9-polarizing conditions (TH9) or TH9 with TL1A (TH9-TL1A) for 3 days. TH9 or TH9-TL1A cells were injected into Rag1−/− mice. a Body weight changes after T-cell transfer. b Histology scores for the intestine. c Representative H&E stains of the duodenum. Scale bar: 200 μm. d Total cell counts in spleens, MLN, and LPMC. e Frequency of CD4+ cells (left). Total cell counts of CD4+ cells (middle, right). f Representative histograms of Ki67 staining of MLN cells (left). Frequency of Ki67+ cells in MLN, spleens, and LPMC (middle). Total cell counts of Ki67+ cells in MLN, spleens, and LPMC (right). g ELISA analysis of IL-9, IL-17, and IL-13 production from MLN after ex vivo restimulation with anti-CD3 and anti-CD28. h ELISA analysis of IL-9 and IL-13 production from LPMC after ex vivo restimulation with anti-CD3 and anti-CD28. i Total cell numbers of IL-9+, IL-13+, and IL-17+ CD4+CD45.1+ T cells in spleens (left) and MLN (right). j Total cell numbers of CD4+CCR6+, and CD4+CD103+ T cells in spleens (left), MLN (middle), and LPMC (right). Data represent means ± SD of one independent experiment out of three independent experiments. n = 4–5/group. *p < 0.05, **p < 0.01, ***p < 0.005 as determined by Student’s t test
IL-9 is required for in vivo inflammation driven by TH9-TL1A cells
To examine whether increased IL-9 production by TH9-TL1A cells contributed to intestinal and lung inflammation, we treated mice receiving TH9-TL1A cells with neutralizing IL-9 or control antibodies for the duration of the T-cell transfer experiment. Histopathological analysis demonstrated significantly reduced intestinal inflammation in anti-IL-9-treated mice compared to isotype controls (Fig. 6a, b). We also observed attenuated lung inflammation with decreased peribronchial infiltrates and reduction in mucus-producing cells in anti-IL-9-treated mice (Supplementary Figure 6c). Total cell numbers and total CD4+CD45.1+ T cells in MLN were reduced in anti-IL-9-treated mice compared to isotype controls (Fig. 6c). Anti-IL-9 treatment also reduced the production of IL-13 and IL-17 in MLN and spleens, while IFN-γ production was unchanged (Fig. 6d, data not shown). Together, these data demonstrate that TH9-TL1A cells were potent effector cells and resulted in intestinal and lung inflammation in vivo, and that the pathogenic effect was IL-9-dependent.
IL-9 is required for in vivo mucosal inflammation driven by TH9-TL1A cells. TH9 or TH9-TL1A cells were injected into Rag1−/− mice and mice were treated three times per week with anti-IL-9 antibody or isotype control. a Representative H&E stainings of the duodenum. Scale bar: 200 μm. b Histology scores for the intestine. c Total MLN cell counts (left), frequency of CD45.1+ cells (middle), and total cell counts of CD45.1+ cells (right). d ELISA analysis of IL-9, IL-17, and IL-13 production from MLN after ex vivo restimulation with anti-CD3 and anti-CD28. Data represent means ± SD of one independent experiment out of two independent experiments. n = 4–5/group. *p < 0.05, **p < 0.01, as determined by Student’s t test (b) or Mann–Whitney U test (c, d)
BATF3 contributes to in vivo inflammation driven by TH9-TL1A cells
To determine whether BATF3 plays a role in TH9-driven pathologies in vivo, we adoptively transferred ex vivo-differentiated WT or Batf3−/− TH9-TL1A into Rag1−/− mice. Mice that received Batf3−/− TH9-TL1A cells develop significantly less pulmonary (Fig. 7a, b, Supplementary Figure 6e) and a trend toward reduced intestinal inflammation (Histoscore: 3.6 vs. 1.2) (Fig. 7a). We observed reduced cellularity in lungs and LPMC, and lower percentages of MLN and lung CD4+ T cells in Batf3−/− TH9-TL1A recipients compared to WT TH9-TL1A recipients (Fig. 7c, Supplementary Figure 6d). Significantly, lower percentages of IL-13+ and IL-17+ cells in spleens, MLN, and lungs from Batf3−/− TH9-TL1A recipients were observed (Fig. 7d). Collectively, our data indicate that BATF3 plays a role in IL-9 production and TH9 development in vitro and in TH9-TL1A-driven mucosal inflammation in vivo.
Batf3 deficiency leads to reduced mucosal inflammation driven by TH9-TL1A cells. Naive WT or Batf3−/− cells were differentiated into TH9-TL1A cells and injected into Rag1−/− mice. a Representative H&E stainings of lungs (top panels) and large intestines (bottom panels). Scale bar: 200 mm. b Histology scores for lungs. c Total cell counts (left, middle), frequency of CD4+ T cells (right). Data represent means ± SD. n = 10/group. d Intracellular staining of IL-9, IL-13, and IL-17 from spleen, MLN, and lungs after ex vivo restimulation with PMA + Ionomycin. Data represent means ± SD. n = 5/group. One representative experiment out of three independent experiments is shown. *p < 0.05, **p < 0.01 as determined by Student’s t test
Discussion
TH9 cells are important for the development of inflammatory and allergic diseases. However, the transcriptional program and inflammatory triggers that drive the differentiation of TH9 cells remain incompletely understood. In this study, we identified the pro-inflammatory cytokine TL1A as a potent inducer of murine and human TH9 differentiation and identified BATF3 as an important transcription factor involved in the differentiation of TH9 cells. The transcriptional network that was induced by TL1A included the transcription factors BATF and BATF3 and several BATF-regulated genes (Supplementary Figure 7). In vivo, in adoptive transfer experiments, TH9-TL1A cells were highly pro-inflammatory leading to intestinal and lung inflammation. The pro-inflammatory propensity of TH9-TL1A cells was dependent on IL-9 production. Transfers of Batf3−/− TH9-TL1A cells resulted in reduced mucosal inflammation and cytokine expression in vivo.
Although TH9 cells have been identified as a distinct T-helper subset, a transcription factor that acts as a master regulator or solely identifies the TH9 phenotype has not been identified. Instead, several transcription factors act in concert to regulate the differentiation of TH9 cells. BATF has been shown to functionally cooperate with IRF4 and to bind to composite elements within the promoter regions of responsive genes. BATF along with IRF4 have been recently proposed as “pioneer factors” in the differentiation of TH17 cells by contributing to the initial chromatin accessibility and facilitating the access of other transcription factors.36 We observed that TL1A has a direct effect on BATF expression and it synergizes with TGF-β1 and IL-4 to maximally induce BATF expression particularly early in TH9 differentiation. In support of this notion, stimulation with TL1A alone leads to the binding of BATF to the Il9 promoter while it has no direct effect on the binding of other transcription factors (IRF4 and BATF3). However, TH9-TL1A conditions synergistically enhance the binding of BATF3 and IRF4, as well as acetylation of histone H3, a permissive chromatin modification that correlates with Il9 promoter activity.11,14 In line with previously published data, BATF is required for the expression of IL-9 and IL-10 under TH9 conditions.15 TL1A is at least partially able to overcome the requirement for BATF in the induction of IL-9 and IL-13 most likely through enhancement of other transcription factors that might be able to compensate the loss of BATF. One candidate for functional compensation is BATF3 which is transcriptionally induced by TL1A. It has been proposed that BATF and BATF3 are functionally interchangeable for TH17 and TH2 development and overexpression of BATF3 in Batf−/− T cells can restore IL-17 production in TH17 cells and IL-4 and IL-10 production in TH2 cells.18,32,37,38 However, the role of BATF3 in the development of TH cells under physiological conditions and potential interaction between BATF and BATF3 during TH9 differentiation has not been defined. We demonstrate here that TL1A facilitates the binding of BATF3 to the Il9 promoter. As a functional consequence, BATF3 contributes to IL-9 production under TH9 conditions particularly in the context of TL1A stimulation. Surprisingly, BATF3 is dispensable for the early differentiation of TH9 cells but might be required to stabilize or maintain the TH9 phenotype and IL-9 secretion at later time points. Our findings of a role of BATF3 in TH9 differentiation are in contrast with previous reports of BATF3 being dispensable for TH1, TH2, and TH17 differentiation.21,37 In contrast to BATF, BATF3 only contributes to IL-9 secretion and not to other TH9 cytokines such as IL-10 and IL-13, suggesting that BATF3 specifically binds to the Il9 promoter and induces IL-9 secretion. Further studies are required to determine additional BATF3 target genes during TH9 differentiation and its role in other TH subsets.
Although other pro-inflammatory cytokines or mediators such as IL-1β, OX40, and TSLP have been described to enhance TH9 differentiation, TL1A seems to be unique in upregulating BATF, thereby opening chromatin for the recruitment of other transcription factors including BATF3.35,39,40 Our RNA-sequencing data support this conclusion identifying a subset of BATF-dependent genes significantly upregulated by TL1A. Furthermore, our RNA-sequencing data reveal that BATF3 regulates the activity of transcription factors, cell proliferation, and intracellular signal transduction suggesting an important role of BATF3 during TH9 differentiation. In vivo, Batf3−/− TH9-TL1A cells resulted in reduced mucosal inflammation and cytokine expression. We did not observe impaired migration of Batf3−/− TH9-TL1A cells to the intestine, which is in contrast to Batf−/− CD4+ T cells that fail to upregulate intestinal migration markers and do not populate the intestines.41 Our data support a role of BATF3 in the pathophysiological function of TH9-TL1A cells.
IL-9 and TH9 cells have been recently associated with the pathogenesis of IBD.6,7,8,9,10 IL-9+ CD4+ T cells were enriched in patients with UC and high serum levels of IL-9 correlated with severe disease in patients with CD.6,7,8,9,10 In an experimental model of hapten-induced UC-like disease, IL-9- and PU.1-deficient mice are protected from intestinal inflammation and anti-IL-9 antibodies are protective in a chronic prevention model.6 However, we did not observe any effects of TL1A on PU.1 expression under TH9 conditions. We have shown that TL1A stimulation results in upregulation of BATF and BATF3 but not of PU.1. Although PU.1 has been described as a master regulator of TH9 differentiation, it is only required for the expression of a subset of TH9 genes and binding of PU.1 is not changed in Batf–/– mice suggesting a complete independency of PU.1 and BATF.15
We demonstrate that TH9-TL1A cells are highly pathogenic in vivo and induce intestinal and lung inflammation. Intestinal inflammation in TH9-TL1A recipients was mainly confined to the small intestine. Furthermore, we observed an increased number of CCR6+ and CD103+ cells in TH9-TL1A recipients in vivo. These data are consistent with findings demonstrating that the chemokine receptor CCR6 and the integrin CD103 are expressed on TH9 cells and facilitate migration to inflammatory sites and mucosal surfaces.15,42 The increased number of CCR6+ cells in TH9-TL1A recipients in vivo is consistent with our observation of mainly small intestinal inflammation since the CCR6/CCL20 axis has been shown to specifically facilitate the migration of TH17 cells to the small intestine and it is plausible that similar mechanisms apply to TH9 cells.43 Our findings are also consistent with recent findings of elevated IL-9 serum levels in CD patients, particularly with severe disease.8
TL1A enhances TH1, TH2, and TH17 differentiation and although we did not see any indication of a global shift from TH9 cells into other TH subsets in vitro, it could suggest that TH9 cells differentiated with TL1A might be unstable in vivo and “trans-differentiate” into other subsets. The potential of trans-differentiation of TH9 cells has been controversial and might be dependent on the context, disease model, and immune status of the host. In EAE, trans-differentiation of TH9 to IFN-γ-producing cells occurs, however, the TH9 phenotype seems to be stable in cancer models.3,39 Surprisingly, in vivo TH9-TL1A cells continued to produce IL-9 and downstream cytokines such as IL-17 and IL-13 and did not trans-differentiate. This notion is also supported by our data showing that anti-IL-9 treatment alone is sufficient to block the pathogenic effects of TH9-TL1A cells. Anti-IL-9 antibody treatment alone reduced IL-13 production and attenuated intestinal and lung inflammation back to TH9 levels. Development of allergic lung inflammation requires the corporation of TH2 and TH9 cells. However, our model of chronic, nonallergic lung inflammation seems to be mainly driven by IL-9-producing TH9 cells, although we cannot exclude a contribution of IL-9-producing non-T cells in pathogenesis in our model.
In summary, TL1A is a strong inducer of murine and human TH9 differentiation and we elucidated the signaling pathways involved. Given the pro-inflammatory properties of TH9-TL1A cells in vivo, targeting the TL1A-BATF3-IL-9 axis may be a promising therapeutic approach in TH9-driven pathologies such as in IBD or allergic lung disease.
Methods
Mice
C57BL/6J, C57BL/6 Rag1−/−, B6.SJL-Ptprca Pepcb/BoyJ (CD45.1), Stat6−/− (B6.129S2(C)-Stat6tm1Gru/J), p50−/− (B6.Cg-Nfkb1tm1Bal/J), B6.Cg-Tg(TcraTcrb)425Cbn/J (OT-II), Batf−/− (B6.129S-Batftm1.1kmm/J), and Batf3−/− (B6.129 S(C)-Batf3tm1kmm/J) mice were purchased from the Jackson Laboratory. Dr3−/− mice have been described elsewhere.44 Mice were maintained under SPF conditions. All animal studies were approved by the Cedars-Sinai Medical Center Animal Care and Use Committee.
T-cell isolation and differentiation
Naive T cells (CD62LhighCD44lowCD25−; clones MEL-14, IM7, PC61.5; eBioscience) were isolated from spleens and MLN using the EasySepTM Mouse CD4+ Isolation Kit (Stem Cell Technologies) followed by cell sorting (MoFlo™, Beckman Coulter). Cells were cultured in RPMI-1640 medium (10% FBS) with anti-CD3ε (145-2C11; BD Biosciences), anti-CD28 (37.51; eBioscience) (TH0 conditions), and murine TL1A (100 ng/ml; R&D Systems) under TH9 conditions (human TGF-β1 [3 ng/ml], BioLegend, IL-4 [20 ng/ml], PeproTech). After 2 days, IL-2 (20 U/ml) was added. To evaluate cell proliferation, sorted cells were labeled with carboxy fluorescein succinimidyl ester (CFSE; Invitrogen), differentiated for 5 days, and CFSE dilution was assessed by flow cytometry. To evaluate antigen-specific TH9 differentiation, naive CD4+ T cells from OT-II mice were stimulated for 3 days with 1 μg/ml OVA323–339 peptide in the presence of syngeneic APCs (ratio 1:3). APCs were prepared by depletion of CD90.2+ T cells from splenocytes (Stem Cell Technologies) followed by mitomycin C treatment.
Isolation of human CD4+ T cells and differentiation
Blood was obtained from healthy volunteers after informed consent in accordance with procedures established by the Cedars-Sinai Institutional Review Board (IRB # 3358, 2673). Naive CD4+ T cells (CD4+CD25–CD45RA+CD45RO−) were isolated from PBMC using the EasySepTM Human CD4+ T cell Enrichment Kit (Stem Cell Technology) followed by cell sorting. Cells were cultured with anti-CD3 (5 μg/ml) and anti-CD28 (2 μg/ml) with human TL1A (100 ng/ml; Fitzgerald) under TH9 conditions (human TGF-β1 [5 ng/ml], human IL-4 [10 ng/ml]).
ELISA
Cytokine concentration in culture supernatants was assayed by ELISA for murine or human IL-9, IL-10, and IL-13 (eBioscience).
Intracellular staining
Cells were restimulated with 50 ng/ml PMA (phorbol 12-myristate 13-acetate), 500 ng/ml ionomycin, and monensin (eBioscience) for 4 h, stained with anti-CD4 (RM4-5, eBioscience), fixed and permeabilized using the FoxP3 staining buffer set (eBioscience), and stained with antibodies against murine IL-9 (RM9A4, BioLegend), IL-10 (JES5-16E3), IL-13 (13A), IL-17A (eBio17B7), IL-17F (eBio18F10), IFN-γ (XMG1.2), IL-4 (BVD6-24G2), IL-22 (1H8PWSR), Ki67 (SolA15), BATF (MBM7C7), IRF4 (3E4), human IL-9 (MH9A4, all eBioscience), and BATF3 (841792, R&D Systems). Samples were analyzed using a CyAn™ ADP flow cytometer (Dako Cytomation) and FlowJo software (TreeStar Inc.).
Quantitative RT-PCR
Total RNA was isolated using RNeasy kits and reverse transcribed into cDNA with Omniscript RT kit (both Qiagen). QPCR was performed using the Mastercycler® ep realplex2 System (Eppendorf). Platimum® Quantitative PCR SuperMix-UDG (Invitrogen) and TaqMan probes and primers were used for Actb, Il9, Il10, Il13, and Batf (IDT) (Supplementary Table 7). RT2 SYBR® Green qPCR Mastermix (Qiagen) and primer sets were used for Actb and Batf3 (IDT). mRNA expression of target genes was normalized to the expression of Actb. The relative gene expression was calculated by the 2−ΔΔCt method.
Chromatin immunoprecipitation
ChIP was performed using EZ ChIP chromatin immunoprecipitation kit (Millipore) followed by qPCR analysis. Totally, 1 × 106 cells were stimulated, fixed, sonicated, and immunoprecipitated using 2 μg of ChIP-grade antibodies: anti-BATF (sc-100974), anti-BATF3 (sc-162246), normal mouse IgG (sc-2025), anti-IRF4 (sc-6059), normal goat IgG (sc-2028) (all Santa Cruz Biotechnology), anti-acetyl-histone H3 (17-615), and normal rabbit IgG (Millipore). Immunoprecipitated DNA was reverse cross-linked, purified using spin columns, and analyzed by qPCR (primer sequences: Supplementary Table 7). To quantify immunoprecipitated DNA, we generated a standard curve from serial dilutions of input DNA. Data are presented as the percentage of input DNA based on normalization against the amount of input DNA.
RNA sequencing (GEO accession numbers: GSE60362 and GSE106926)
Low-input RNA-Seq (TH9 vs. TH9-TL1A): Clontech’s SMARTer® Ultra™ Low Input RNA Kit for Sequencing v3 was employed to generate double-stranded cDNA libraries from 1.5 to 2.5 ng of total RNA from each sample as per the manufacturer’s recommendations. Double-stranded cDNA libraries were individually fragmented and ligated with Ion Torrent Ion Xpress™ Barcode Adapters using Ion Xpress™ Plus Fragment Library Kit with limited modifications including size-selected final libraries with a double-bead cleanup and ½ volume of ion adapters. RNA-Seq libraries were assessed for concentration and length using Invitrogen’s Qubit® dsDNA HS Assay Kit and Agilent High Sensitivity DNA Kit, respectively. Samples were multiplexed to obtain >10 million reads each for sequencing. The pooled libraries were amplified onto Ion Sphere™ and sequenced using the Ion PI™ Sequencing 200 Kit v2, Ion Torrent™.
mRNA-Seq (WT vs. Baft3−/− TH9-TL1A): Illumina TruSeq Stranded mRNA library preparation kit was employed for mRNASeq. Of total RNA, 1 μg per sample was used for poly-A mRNA selection using streptavidin-coated magnetic beads. cDNA was synthesized from enriched and fragmented RNA using reverse transcriptase (Super-Script II, Invitrogen) and random primers. The cDNA was converted into double-stranded DNA, and enriched with PCR for library preparation. The PCR-amplified library was purified using Agencourt AMPure XP beads (Beckman Coulter). The concentration of the amplified library was measured with a NanoDrop spectrophotometer and on an Agilent 2100 Bioanalyzer. Samples were multiplexed to obtain >20 million reads/sample and sequenced on a NextSeq 500 platform (Illumina) using 75-bp single-end sequencing.
Data analysis
Raw reads were filtered and trimmed by FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and aligned using Tophat version 2.0.8 with UCSC GRCm38/mm10 mouse reference genome annotation (http://genome.ucsc.edu). Gene read counts were generated using HTSeq (v0.5.4) and then normalized by the trimmed mean of the M-values normalization method with edgeR (v3.0.8) in Bioconductor in R (v2.15), which uses a weighted trimmed mean of the log expression ratios. For all analysis, only quality signals (threshold: ten counts per million in at least two out of four samples) were used. Unsupervised analysis was performed using the top 100 ranked genes by interquartile range and then hierarchical clustering was generated using gplots (v2.11) using two-way Pearson correlations to visualize unbiased pervasive gene expression patterns. Supervised analysis using a modified Fisher’s exact test in edgeR and a false discovery rate cutoff of 10% using Benjamini and Hochberg procedure was used to determine differential expressions between TH9 and TH9-TL1A or WT and Batf3−/− TH9-TL1A. Pathway enrichment analysis was performed using DAVID 6.7 (http://david.abcc.ncifcrf.gov/) with count threshold = 5 and ease score threshold = 0.05.
T-cell transfer model
CD4+CD62LhighCD44lowCD25− T cells from CD45.1 mice were differentiated into TH9 or TH9-TL1A cells for 3 days. Male Rag1−/− recipient mice were injected i.p. with 0.5 × 106 TH9, or TH9-TL1A cells. Mice were weighed for 6–8 weeks. For neutralization experiments, mice were injected i.p. with anti-IL-9 or IgG2b control antibody (both 50 μg/dose; R&D Systems) three times per week for 4 weeks and twice per week for the remaining time starting at the day of T-cell transfer. Tissues were formalin-fixed, paraffin-embedded, and stained with H&E or AB-PAS. Inflammation was scored by a semiquantitative scoring system by a trained pathologist blinded to the experimental conditions.45,46 LPMC were isolated from the large intestine as previously described.47 Lung tissue was digested for 45 min at 37 °C with 10 ml of HBSS containing 15 μg/ml Liberase™ (Roche) and 25 μg/ml DNase I (Sigma) and filtering through a 40-μm cell strainer followed by red blood cell lysis. Single-cell suspensions were stained with anti-CD4, anti-CD45.1 (A20), anti-CCR6 (29-2L17), anti-CD103 (2E7, all eBioscience), and anti-Ki-67 (SolA15, BD PharMingen) antibodies for flow cytometry or restimulated with anti-CD3ε/anti-CD28 antibodies for 3 days. Cytokine levels in supernatants were measured by ELISA.
Statistics
Statistical significance was calculated using SPSS software (IBM SPSS Statistics 20, IBM Machines Corp., Armonk, NY, USA). Data were analyzed using one-way analysis of variance (ANOVA) followed by post hoc unpaired two-tailed Student t test, or Mann–Whitney U test as indicated. Differences were considered significant at p < 0.05.
References
Dardalhon, V. et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(−) effector T cells. Nat. Immunol. 9, 1347–1355 (2008).
Veldhoen, M. et al. Transforming growth factor-beta “reprograms” the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 9, 1341–1346 (2008).
Jager, A., Dardalhon, V., Sobel, R. A., Bettelli, E. & Kuchroo, V. K. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J. Immunol. 183, 7169–7177 (2009).
Licona-Limon, P. et al. Th9 cells drive host immunity against gastrointestinal worm infection. Immunity 39, 744–757 (2013).
Purwar, R. et al. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat. Med. 18, 1248–1253 (2012).
Gerlach, K. et al. TH9 cells that express the transcription factor PU.1 drive T cell-mediated colitis via IL-9 receptor signaling in intestinal epithelial cells. Nat. Immunol. 15, 676–686 (2014).
Nalleweg, N. et al. IL-9 and its receptor are predominantly involved in the pathogenesis of UC. Gut. 64, 743–755 (2015).
Feng, T. et al. Serum interleukin 9 levels predict disease severity and the clinical efficacy of infliximab in patients with Crohn’s disease. Inflamm. Bowel Dis. 23, 1817–1824 (2017).
Matusiewicz, M., Neubauer, K., Bednarz-Misa, I., Gorska, S. & Krzystek-Korpacka, M. Systemic interleukin-9 in inflammatory bowel disease: association with mucosal healing in ulcerative colitis. World J. Gastroenterol. 23, 4039–4046 (2017).
Defendenti, C. et al. Significance of serum Il-9 levels in inflammatory bowel disease. Int. J. Immunopathol. Pharmacol. 28, 569–575 (2015).
Jash, A. et al. Nuclear factor of activated T cells 1 (NFAT1)-induced permissive chromatin modification facilitates nuclear factor-kappaB (NF-kappaB)-mediated interleukin-9 (IL-9) transactivation. J. Biol. Chem. 287, 15445–15457 (2012).
Staudt, V. et al. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity 33, 192–202 (2010).
Goswami, R. et al. STAT6-dependent regulation of Th9 development. J. Immunol. 188, 968–975 (2012).
Chang, H. C. et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat. Immunol. 11, 527–534 (2010).
Jabeen, R. et al. Th9 cell development requires a BATF-regulated transcriptional network. J. Clin. Invest. 123, 4641–4653 (2013).
Kaplan, M. H. The transcription factor network in Th9 cells. Semin. Immunopathol. 39, 11–20 (2017).
Echlin, D. R., Tae, H. J., Mitin, N. & Taparowsky, E. J. B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene 19, 1752–1763 (2000).
Murphy, T. L., Tussiwand, R. & Murphy, K. M. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 13, 499–509 (2013).
Sopel, N., Graser, A., Mousset, S. & Finotto, S. The transcription factor BATF modulates cytokine-mediated responses in T cells. Cytokine Growth Factor Rev. 30, 39–45 (2016).
Edelson, B. T. et al. Peripheral CD103(+) dendritic cells form a unified subset developmentally related to CD8 alpha(+) conventional dendritic cells. J. Exp. Med. 207, 823–836 (2010).
Hildner, K. et al. Batf3 deficiency reveals a critical role for CD8alpha + dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008).
Meylan, F. et al. The TNF-family receptor DR3 is essential for diverse T cell-mediated inflammatory diseases. Immunity 29, 79–89 (2008).
Fang, L., Adkins, B., Deyev, V. & Podack, E. R. Essential role of TNF receptor superfamily 25 (TNFRSF25) in the development of allergic lung inflammation. J. Exp. Med. 205, 1037–1048 (2008).
Pappu, B. P. et al. TL1A-DR3 interaction regulates Th17 cell function and Th17-mediated autoimmune disease. J. Exp. Med. 205, 1049–1062 (2008).
Bull, M. J. et al. The death receptor 3-TNF-like protein 1A pathway drives adverse bone pathology in inflammatory arthritis. J. Exp. Med. 205, 2457–2464 (2008).
Thomas, L. S. et al. The TNF family member TL1A induces IL-22 secretion in committed human Th17 cells via IL-9 induction. J. Leukoc. Biol. 101, 727–737 (2017).
Richard, A. C. et al. The TNF-family ligand TL1A and its receptor DR3 promote T cell-mediated allergic immunopathology by enhancing differentiation and pathogenicity of IL-9-producing T cells. J. Immunol. 194, 3567–3582 (2015).
Tan, C. et al. Antigen-specific Th9 cells exhibit uniqueness in their kinetics of cytokine production and short retention at the inflammatory site. J. Immunol. 185, 6795–6801 (2010).
Wilhelm, C. et al. An IL-9 fate reporter demonstrates the induction of an innate IL-9 response in lung inflammation. Nat. Immunol. 12, 1071–1077 (2011).
Bamias, G. et al. Role of TL1A and its receptor DR3 in two models of chronic murine ileitis. Proc. Natl Acad. Sci. USA 103, 8441–8446 (2006).
Betz, B. C. et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 207, 933–942 (2010).
Schraml, B. U. et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 460, 405–409 (2009).
Glasmacher, E. et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 338, 975–980 (2012).
Li, P. et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature 490, 543–546 (2012).
Yao, W. et al. Interleukin-9 is required for allergic airway inflammation mediated by the cytokine TSLP. Immunity 38, 360–372 (2013).
Ciofani, M. et al. A validated regulatory network for Th17 cell specification. Cell 151, 289–303 (2012).
Tussiwand, R. et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 490, 502–507 (2012).
Iwata, A. et al. Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF-IRF4 transcription factor complex. Nat. Immunol. 18, 563–572 (2017).
Vegran, F. et al. The transcription factor IRF1 dictates the IL-21-dependent anticancer functions of TH9 cells. Nat. Immunol. 15, 758–766 (2014).
Xiao, X. et al. OX40 signaling favors the induction of T(H)9 cells and airway inflammation. Nat. Immunol. 13, 981–990 (2012).
Wang, C. et al. BATF is required for normal expression of gut-homing receptors by T helper cells in response to retinoic acid. J. Exp. Med. 210, 475–489 (2013).
Kara, E. E. et al. Distinct chemokine receptor axes regulate Th9 cell trafficking to allergic and autoimmune inflammatory sites. J. Immunol. 191, 1110–1117 (2013).
Esplugues, E. et al. Control of TH17 cells occurs in the small intestine. Nature 475, 514–518 (2011).
Shih, D. Q. et al. Inhibition of a novel fibrogenic factor Tl1a reverses established colonic fibrosis. Mucosal Immunol. 7, 1492–1503 (2014).
Ostanin, D. V. et al. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G135–G146 (2009).
Chin, J. E. et al. Airway recruitment of leukocytes in mice is dependent on alpha4-integrins and vascular cell adhesion molecule-1. Am. J. Physiol. 272, L219–L229 (1997).
Weigmann, B. et al. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat. Protoc. 2, 2307–2311 (2007).
Acknowledgements
This work was supported by the NIH (DK056328 to S.R.T.) and the F. Widjaja Foundation (S.R.T. and K.S.M.). Anita Vibsig Neutzsky-Wulff received postdoctoral fellowships from The Carlsberg Foundation (Denmark) and the Lundbeck Foundation (Denmark). Jordan Nunnelee received a Student Research Award by the Crohn’s and Colitis Foundation of America. The Cedars-Sinai MIRIAD IBD Biobank is supported by the F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute, NIH/NIDDK grants P01 DK046763, U01 DK062413, and The Leona M and Harry B Helmsley Charitable Trust.
Author information
Authors and Affiliations
Contributions
M.T., H.H., L.T., M.W., B.S., M.W., J.N., J.S., A.N.-W., R.B., Y.W., J.T., V.F., and K.M. performed experiments, analyzed data, and critically reviewed the manuscript. A.P., J.T., and Y.W. performed RNA-Seq data analysis. M.T., S.T., and K.M. designed the experiments. M.T. and K.M. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Tsuda, M., Hamade, H., Thomas, L.S. et al. A role for BATF3 in TH9 differentiation and T-cell-driven mucosal pathologies. Mucosal Immunol 12, 644–655 (2019). https://doi.org/10.1038/s41385-018-0122-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-018-0122-4
This article is cited by
-
Concise review: The heterogenous roles of BATF3 in cancer oncogenesis and dendritic cells and T cells differentiation and function considering the importance of BATF3-dependent dendritic cells
Immunogenetics (2024)
-
IL-9-producing T cells: potential players in allergy and cancer
Nature Reviews Immunology (2021)
-
Intestinal epithelium-derived BATF3 promotes colitis-associated colon cancer through facilitating CXCL5-mediated neutrophils recruitment
Mucosal Immunology (2021)
-
What's old is new again: Batf transcription factors and Th9 cells
Mucosal Immunology (2019)