Abstract
The lung is the primary site of infection with the major human pathogen, Mycobacterium tuberculosis. Effective vaccines against M. tuberculosis must stimulate memory T cells to provide early protection in the lung. Recently, tissue-resident memory T cells (TRM) were found to be phenotypically and transcriptional distinct from circulating memory T cells. Here, we identified M. tuberculosis-specific CD4+ T cells induced by recombinant influenza A viruses (rIAV) vaccines expressing M. tuberculosis peptides that persisted in the lung parenchyma with the phenotypic and transcriptional characteristics of TRMs. To determine if these rIAV-induced CD4+ TRM were protective independent of circulating memory T cells, mice previously immunized with the rIAV vaccine were treated with the sphingosine-1-phosphate receptor modulator, FTY720, prior to and during the first 17 days of M. tuberculosis challenge. This markedly reduced circulating T cells, but had no effect on the frequency of M. tuberculosis-specific CD4+ TRMs in the lung parenchyma or their cytokine response to infection. Importantly, mice immunized with the rIAV vaccine were protected against M. tuberculosis infection even when circulating T cells were profoundly depleted by the treatment. Therefore, pulmonary immunization with the rIAV vaccine stimulates lung-resident CD4+ memory T cells that are associated with early protection against tuberculosis infection.
Similar content being viewed by others
Introduction
Tuberculosis (TB) is the major cause of death from a bacterial pathogen in adults worldwide; in 2016 alone there were 1.6 million deaths and an estimated 10.4 million new cases.1 The current vaccine, BCG, although able to protect children from disseminated infection, fails to protect the adults from the contagious pulmonary form of disease.2 The protective immune response against M. tuberculosis is mostly dependent on CD4+ T cells3 as evidenced by the increased susceptibility of individuals with HIV/AIDS to TB.4 Novel TB vaccines that induce T cells are being actively pursued, but so far with limited success,5 suggesting that new strategies are required
Until recently, memory T cells were considered to exist as two main subsets that exclusively circulated from the blood into tissues or lymphoid organs. While central memory T cells (TCM) are able to enter lymphoid organs, effector memory T cells (TEM) circulate through peripheral sites. The observation that subsets of virus-specific long-lived CD8+ T memory cells were located in nonlymphoid tissues6 suggested the existence of non-circulating populations of tissue-resident memory T cells (TRM). Their tissue residence was subsequently confirmed by parabiosis studies.7
TRM found in several tissues, including the lung,6 are phenotypically and functionally distinct from their circulating TCM and TEM memory T-cell counterparts. They express low levels of the transcription factor, Kruppel-like factor 2 (KLF2), and spinghosine-1-phosphate (S1P) receptor, S1PR1, and high levels of C-type lectin, CD69.8 Expression of S1PR1 and its regulatory transcription factor KLF2 are essential for promoting T lymphocyte egress into circulation.9,10 Downregulation of these two genes in TRM prevent them from sensing the S1P gradient in the circulation,11 thus inhibiting their exit from tissues.
The presence of TRM in tissues has been associated with protection against several pathogens, and the high number of TRM found in mucosal sites support the concept that these cells represent a first line of defense against pathogens.8 Most studies have focused on CD8+ TRM cells in the context of viral infections and demonstrated these cells can protect mice against viruses,12,13,14 Listeria monocytogenes15 and malaria.16 The induction of CD4+ TRM cells has been less studied but growing evidence of the presence and protective function of these cells, particularly in the lung, is emerging.17,18,19
We have previously shown that pulmonary immunization of mice with a viral TB vaccine, recombinant influenza A virus (rIAV) expressing the p25 CD4+ T-cell immunodominant epitope of M. tuberculosis Ag85B (PR8.p25), induced strong p25-specific CD4+ T-cell responses in the lungs that were protective against M. tuberculosis challenge.20 In this study, we have investigated whether pulmonary immunization of mice with rIAVs expressing this p25 epitope induces M. tuberculosis-specific CD4+ T cells in the lung with the tissue location, phenotype, and transcriptional characteristics of TRMs, and if these lung-resident T cells are protective against M. tuberculosis challenge in the absence of circulating memory T cells.
Results
Pulmonary, but not systemic, immunization with PR8.p25 induces P25 CD4+ T cells in the lung
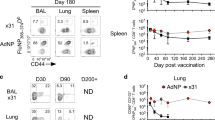
To determine if mucosal immunization with PR8.p25 is required for the induction of the P25-specific CD4+ T-cell response in the lungs, 5 × 104 CD45.1+ P25 cells, expressing a transgenic TCR recognizing the p25 CD4+ T-cell epitope of the M. tuberculosis Ag85B, were transferred intravenously (i.v.) into C57BL/6 mice that were infected on the following day, either intranasally with 20 pfu or intraperitoneally (i.p.) with 104 pfu of PR8.p25. Three weeks later, the P25-specific response was measured in the lung and spleen by interferon (IFN)γ ELISpot and flow cytometry. Intranasal infection with PR8.p25 led to strong responses to both the M. tuberculosis-specific p25 epitope Ag85B240–254 and the IAV endogenous CD8+ T-cell epitope, NP366–374, in both the lung (Fig. 1a) and spleen (Fig. 1b). Intraperitoneal infection with PR8.p25 induced splenic T-cell responses that were comparable to those induced by intranasal immunization, but failed to induce substantial responses in the lungs. These findings were confirmed by flow cytometry as significant numbers of P25 transgenic cells were present in the broncho-alveolar lavage (BAL) and total lung cell suspensions only when mice were infected intranasally with PR8.p25 (Fig. 1c, d). By contrast, similar numbers of P25 donor T cells were found in the spleen after infection by either route (Fig. 1e). These results demonstrate that optimal generation of a p25-specific CD4+ T-cell response in the lungs requires the pulmonary delivery of the vaccine.
Pulmonary, but not systemic, immunization with PR8.p25 induces p25 CD4+ T cells in the lung. Fifty-thousand CD45.1+ P25 Tg splenic cells were adoptively transferred into naive recipient C57Bl/6 1 day prior to immunization intranasally with 20 pfu or intraperitoneally with 5 × 104 pfu of PR8.p25. IFNγ responses in the lung (a) and in the spleen (b) against influenza A virus NP and M. tuberculosis Ag85B P25 epitopes, 3 weeks after delivery of PR8.p25 intraperitoneally (black) or intranasally (gray) measured by IFNγ ELISPOT. Number of P25 CD4+ T cells in the BAL (c), lung (d), and spleen (e) determined by flow cytometry. Data presented as mean ± SEM (n = 4)
Intranasal immunization with PR8.p25 leads to the persistence of CD69+CD11a+ CD44hiCD62Llo P25 CD4+ T cells in the lung
The kinetics of the total CD4+ T cell (Fig. 2a) and P25 T cell (Fig. 2b) responses were monitored after adoptive transfer of P25 cells and intranasal immunization with PR8.p25. The p25-specific CD4+ T-cells response in the lungs underwent a robust expansion 2 weeks after immunization. At 6 weeks post immunization after contraction of the response, a significant number of p25-specific cells (2 × 104) was detected in the lungs (Fig. 2b). The majority of these persisting P25 CD4+ T cells expressed CD69, but less than 5% expressed CD103 (Fig. 2c). Of note, the proportion of P25 CD4+ T cells expressing CD69 increased following infection and by week 8 post infection >90% of the P25 CD4+ T cells in the lung were CD69+ (Fig. 2d). CD69 expression in the persistent P25 CD4+ T cells was not due to recent activation caused by antigen remaining in the lungs. This was confirmed by the failure of CFSE-labeled P25 transgenic CD45.1 + CD4+ T cells to proliferate in the lungs and lymph nodes when adoptively transferred into mice 6 weeks after immunization with PR8.p25. By contrast, CFSE-labeled P25 T cells proliferated when transferred 5 days after immunization (Supplementary Fig S3).
Intranasal immunization with PR8.p25 induces the persistence of CD69+ CD11a+ p25 CD4+ T cells in the lung parenchyma. Fifty-thousand CD45.1+ P25 or GFP-P25 Tg splenic cells were adoptively transferred into naive recipient C57Bl/6 mice 1 day prior to immunization intranasally with 20 pfu PR8.p25. Kinetics of the total CD4 + T cells (a) and of the transferred CD45.1+ P25 CD4+ T cells (b) in the lung after immunization with PR8.p25. c Expression of CD69 and CD103 by the P25 CD4 T cells. d Intravascular staining of the CD3+CD4+ population showing the CD45-unlabeled GFP-P25 T cells located in the lung parenchyma (quadrant I) and the vascular CD45-labeled GFP-P25 T cells (quadrant II), e CD69 expression profile in parenchymal (blue) vs. vascular (red) GFP-P25 T cells, f expression of CD69 and CD11a and g CD44 and CD62L in GFP-P25 parenchymal cells, and h numbers of parenchymal GFP-P25 cells expressing CD69 and CD11a at 8 weeks after immunization with PR8.p25. The data are representative of 1 of 2 independent experiments and are presented as mean ± SEM (n = 3–5)
To assess whether P25 CD4+ T cells persisting in the lung at 8 weeks after immunization were located in the tissue or in the vasculature, mice were injected i.v. with anti-CD45 antibodies 3 min prior to harvesting of the lungs as described.21 The vast majority ( > 95%) of the lung P25 CD4+ T cells failed to be stained by anti-CD45 antibodies, indicating they were located in the lung parenchyma (Fig. 2d). Parenchymal, CD45-unlabeled P25 CD4+ T cells expressed CD69, in contrast to circulating, CD45-labeled P25 CD4+ T cells that did not express this marker (Fig. 2e), Similar low levels of CD69 expression were observed in CD45-labeled CD3+CD4+ T cells present in the lung vasculature (Supplementary Fig S1). Further, phenotypic analysis showed that the majority of P25 CD4+ T cells retained in the lung parenchyma at 8 weeks after immunization with PR8.P25 expressed both CD69 and CD11a (Fig. 2f, h) and were CD44hiCD62Llo (Fig. 2g).
P25 CD4+ T cells induced by immunization with PR8-p25 are distributed throughout the lung parenchyma
To determine the in situ localization of the P25 CD4+ T cells following the rIAV immunization, 2-photon microscopy was performed on the lungs collected from immunized mice. Consistent with flow cytometry results, imaging using 2-photon microscopy showed that at 8 weeks after intranasal PR8.p25 infection, P25 T cells expressing GFP (P25-GFP) were readily detectable in the lungs (Fig. 3a). The extravascular location of these P25-GFP cells was confirmed by imaging the lungs of infected mice in which the vessels were labeled with fluorescent wheat germ agglutinin (Fig. 3b and Supplementary video 1). Furthermore, clearing of the lungs with RapiClear1.52 solution allowing deeper imaging of tissue revealed that the P25-GFP cells were scattered throughout the lung, from the pleura to the more central areas near the airways (Fig. 3c and Supplementary video 2). Quantification of the P25-GFP cells present in the lungs after 8 weeks of PR8.p25 immunization using 2-photon randomly acquired z-stacks showed that the density of these cells was 17–224 cells/mm3 of lung, 57–75 times more that what determined by flow cytometry for the lungs of the same mice (Fig. 3d). Strikingly, 36 weeks after pulmonary immunization of PR8.p25, parenchymal CD69+CD11a+ P25 CD4+ T cells were still detected in the lungs of all the mice analyzed, with a mean number of 133 ± 111 ( ± SD, n = 7). Thus, immunization with PR8.p25 leads to long-term persistence of CD62LloCD44hiCD69+CD11a+ P25 CD4+ memory T cells in the lung parenchyma.
Detection of GFP-P25 CD4+ T cell at 6 weeks after immunization with PR8.p25 using 2-photon microscopy. Fifty-thousand GFP-P25 Tg splenic cells were adoptively transferred into naive recipient C57Bl/6 mice 1 day prior to intranasal immunization with 20 pfu PR8.p25. a GFP-P25 cells (green) were detected in an ex vivo lung from mice immunized with PR8.p25 vaccine 6 weeks after immunization. b The extravascular location of the GFP-P25 cells (green) induced by immunization with PR8.p25 was confirmed after injection of WGA-A594 that allows visualization of the vasculature (red). c Detection of GFP-P25 cells (green) deeper into the tissue after clearing of the lung with RapiClear1.52. d Comparison of the frequency of GFP-P25 cells in the lung as determined by flow cytometry and by 2-P microscopy 8 weeks after immunization with PR8.p25
P25 CD4+ T cells induced by PR8.p25 immunization have the transcriptional characteristics of TRMs
To investigate further the properties of the vaccine-induced memory T cells, lung and splenic P25 T cells from mice immunized intranasally with PR8.p25 6 weeks earlier were sorted according to their memory phenotype, and the transcriptional profiles of naive, effector and the different memory P25 cell subsets were analyzed by qPCR. Lung CD69+ P25 cells displayed a distinct reduction in Klf2 and S1pr1 mRNA levels compared to their splenic CD69-CD62Llo counterparts (Fig. 4a). The transcription of FOXO1, an upstream regulator of KLF2,22 was also more markedly downregulated in the lung CD69+ memory T cells than in splenic memory T cells. Lung CD69+ memory P25 cells also expressed higher levels of Tbx21 and Ifng mRNA and higher levels of pro-survival markers, Bcl2 and Bclxl than splenic effector memory CD4 TEM (Fig. 4a, b). Their results demonstrate that the long-lived, pulmonary M. tuberculosis-specific CD4+ T cells induced by pulmonary immunization with PR8.P25 exhibit the transcriptional profile of tissue-resident memory T cells
P25 CD4+ T cells induced by PR8.p25 immunization have the transcriptional profile of tissue-resident memory cells. Fifty-thousand CD45.1+ P25 Tg splenic cells were adoptively transferred into naive recipient C57Bl/6 mice 1 day prior to immunization intranasally with 20 pfu PR8.p25. Lung CD69+ P25 CD4+ T cells (L-RM) and splenic CD69-CD62Llo (S-EM) were sorted from mice immunized with PR8.p25 6 weeks before (n = 12–20). P25 CD4+ effector cells (S-eff) were sorted from the lungs of mice 11 days after immunization with PR8.p25. Quantitative real-time PCR analysis of (a) klf2, s1pr1, foxo1, tbx21, ifng and (b) bcl-2 and bcl-xl mRNA in P25 cells sorted from the spleens and lungs of naive and PR8-p25 immunized mice. The representative figure shows the results from one of two independent experiments
The persistence of P25 CD4+ T cells induced by rIAV immunization in the lung is independent of replenishment of circulating T cells
To test if the rIAV-induced P25 CD4+ T cells residing in the lung parenchyma were dependent on the continuing recruitment of circulating CD4+ T cells, the response to PR8.p25 was examined in mice with lymphopenia following administration of FTY720, a S1P receptor modulator that inhibits egress of lymphocytes from lymph nodes and other secondary lymphoid organs.23,24 Six weeks after immunization with PR8.p25, mice were administered i.p. 1 mg/kg of FTY720 or phosphate buffer saline (PBS) daily for 20 days. The treatment with FTY720 significantly reduced the percentage of circulating CD3+CD4+ T cells both in the peripheral blood and in the lung vasculature, but did not affect the number of P25 CD69+CD4+ T cells in the lung parenchyma (Supplementary Fig S4). This demonstrated that the P25 CD4+ T cells present in the lung parenchyma 6 weeks after PR8.p25 immunization were non-circulating memory T cells that persisted in the tissue without requiring replenishment from by circulating cells. We then used this model to assess the response of rIAV-induced CD4+ TRM cells against challenge with M. tuberculosis. B6 mice were immunized with rIAV X31.p25 vaccine that encodes the same M. tuberculosis p25 epitope and stimulates lung CD4+ TRM cells that protect mice against M. tuberculosis infection26. Three days after FTY720 treatment both immunized and non-immunized mice were challenged by aerosol infection with low dose M. tuberculosis. The CD4+ T responses to infection in FTY720-treated vs. non-treated mice were assessed on 17 and 28 days post infection (dpi) (Fig. 5a). As expected FTY720 treatment led to significant reduction in the number of circulating CD4+ T cells in both immunized and unimmunized mice at 17 dpi (Fig. 5b). Similar effects were also seen in the MLN (Fig. 5c), lung parenchyma (Fig. 5d and Supplementary Fig. S5) and lung vasculature (Fig. 5e and Supplementary Fig. S5). At 28 dpi, 11 days after terminating FTY720 treatment, the number of CD4+ T cells was similar in all experimental groups (Supplementary Fig. S5). There was a small decrease in the number of donor P25 CD4+ T cells present in the lung parenchyma of the treated mice at day 17 dpi (Fig. 5f) suggesting that at this time-point after M. tuberculosis challenge there was recruitment of circulating GFP-p25 cells in to the lung. Importantly, the GFP-P25 population with the TRM signature (CD11a+CD62LloCD69+KLRG-) was still the most abundant subset in the lung parenchyma of the immunized mice (Fig. 5g) and the number of these cells was not affected by FTY720 treatment (Fig. 5h). Thus, the TRM CD4+ T cells induced by rIAV vaccines were maintained in the lungs independently of circulating T cells.
Effect of FTY720 treatment on CD4+ T cells after infection with M. tuberculosis. Fifty-thousand GFP-P25 Tg splenic cells were adoptively transferred into naive recipient C57Bl/6 mice 1 day prior to intranasal immunization with 5 × 104 pfu X31-p25. Six weeks after immunization, both naive and immunized mice were treated with FTY720, 1 mg/kg by i.p. daily for 20 days and challenged with M. tuberculosis at day 3 of treatment. CD4+ T-cell populations were analyzed and compared with those of naive and immunized mice infected with M. tuberculosis but not treated with FTY720. a Diagram of the experimental plan. The number of CD4+ T cells present in 200 µl of blood (b), mediastinal lymph node (c), lung parenchyma (d), and lung vasculature (e) at 17 days post infection. f Number of GFP-P25 Tg CD4+ T cells, g pie charts representing the proportion of the different subset populations of GFP-P25 CD4+ T cells based on the surface markers CD69, CD11a, CD62L and KLRG1 and h number of CD69+CD11a+CD62LloKLRG1- GFP-P25 CD4+ T cells present in the lung parenchyma at day 17 and 28 post M. tuberculosis infection. Results in b–f and h are shown as mean ± SD (n = 5–7). Results in g were obtained using the FlowJo bolean gating tool. The representative figure shows the results from one of two independent experiments. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, ANOVA followed by Tukey post-test correction
The CD4+ TRMs induced by rIAV immunization produce Th1 cytokines in response to M. tuberculosis challenge
The cytokine production in the lungs of the immunized and unimmunized mice, with or without FTY720 treatment, was analyzed at 17 and 28 dpi by ICS after incubation of cells in the presence of p25 peptide. At 17 dpi, both groups of mice immunized with the rIAV vaccine had significantly higher magnitude of CD4+ T cell cytokine responses than the unimmunized groups, particularly triple-cytokine (IFNγ/TNFIL-2) producers and double (IFNγ/TNF) producers (Fig. 6a). The majority of the cytokine-producing T cells was located in the lung parenchyma (Supplementary Fig. S6) and corresponded to donor GFP-P25 CD4+ T cells (Fig. 6b). FTY720 treatment had no effect on the cytokine production by GFP-P25 CD4+ T cells at any time-point (Fig. 6b, c). At 28 dpi, the cytokine profile changed into a more effector phenotype consisting of IFNγ or IFNγ/TNF production by donor GFP-P25 CD4+ T cells (Fig. 6c) and other p25-specific CD4+ T cells without significant differences between groups (Supplementary Fig. S7). Therefore, the TRM CD4+ T cells induced by rIAV vaccines were able to produce macrophage-activating cytokines in response to M. tuberculosis challenge.
CD4+ T-cell cytokine production in M. tuberculosis infected lungs after FTY720 treatment. Fifty-thousand GFP-P25 Tg splenic cells were adoptively transferred into naive recipient C57Bl/6 mice 1 day prior to intranasal immunization with 5 × 104 pfu X31-p25. Naive and mice immunized with X31-p25 were treated with FTY720 and challenged with M. tuberculosis as in Fig. 5. Lung cells were stimulated with p25 peptide and the cytokine production assessed by ICS assay. a Frequency of cytokine-producing CD4+ T cells in the lung parenchyma at 17 days post M. tuberculosis infection. Cytokine production by GFP-P25 CD4+ T cells in the lung parenchyma at 17 days (b) and 28 days (c) post M. tuberculosis infection. Pie charts represent the relative proportions of p25-specific CD4+ T cells expressing the possible combinations of IFNγ, TNF, and IL-2. Analysis of the different cytokine-producing subsets was performed by flow cytometry using FlowJo Bolean gating tool. Results are shown as mean + SD (n ≥ 5) and are from one of two independent experiments. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, ANOVA followed by Tukey post-test correction
The CD4+ TRMs induced by rIAV immunization are associated with protection against M. tuberculosis in the lung
Consistent with the findings of others,25 previous studies in our laboratory have shown that neither simultaneous (data not shown) nor prior infection with wild-type PR8 IAV (Supplementary Fig. S8) protect C57BL/6 mice against M. tuberculosis infection. Similar lack of protection was found after immunization with another rIAV vaccine (PR8-TB10.4) expressing a M. tuberculosis-specific CD8+ T-cell epitope20 suggesting that the protection conferred by PR8.p25 is due to the P25 T cells generated and not caused by non-specific responses to the rIAV. In addition, immunization with wild-type X31 or PR8 IAVs do not induce endogenous p25-specific CD4+ T-cell responses20,26 and the wild-type X31 virus does not induce the expansion of transferred p25 tg T cells.26
To determine if the CD4+ TRMs induced by the rIAV vaccination protected mice against M. tuberculosis, the mycobacterial loads contained in the lungs of immunized and non-immunized mice with or without FTY720 treatment were determined at 17 and 28 days after aerosol challenge (Fig. 5a). At 17 dpi, rIAV vaccination conferred significant protection in the lung. More importantly, the vaccine-induced responses were protective in FTY720-treated mice (Fig. 7a), in which the circulating CD4+ T cells were substantially depleted (Fig. 5a). At 28 dpi, unimmunized mice treated with FTY720 showed a trend towards higher bacterial loads compared to the non-treated unimmunized mice (Fig. 7b). Although there were no differences in the number of CD4+ T cells at 28 dpi between groups (Supplementary Fig. S6) this enhanced susceptibility to infection may be due to a delay in the recruitment of cells to the lung caused by the FTY720 treatment. Strikingly, this effect was absent in mice that were immunized with the rIAV vaccine (Fig. 7b), strongly suggesting that the presence of lung TRMs induced by the rIAV vaccine not only conferred protection, but also limited the increased susceptibility to M. tuberculosis infection induced by prior FTY720 treatment.
X31.p25-induced lung-resident CD4+ T cells confer protection against M. tuberculosis following depletion of circulating T cells. Fifty-thousand GFP-P25 Tg splenic cells were adoptively transferred into naive recipient C57Bl/6 mice 1 day prior to intranasal immunization with 5 × 104 pfu X31-p25. Naive and mice immunized with X31-p25 were treated with FTY720 and challenged with M. tuberculosis as in Fig. 5. Mycobacterial loads in the lungs of mice were determined at 17 (a) and 28 days (b) after infection with M. tuberculosis. The data are shown with symbols for individual animals and as mean ± SEM (n = 6) and are representative of results from one of two independent experiments. ****p < 0.0001,***p < 0.001, **p < 0.01, ANOVA followed by Tukey post-test correction
Discussion
There is growing interest in developing vaccines that are able to induce tissue-resident memory cells at the site of subsequent infection. This is particularly relevant for TB vaccines since direct recognition of infected macrophages by CD4+ T cells is required for protection against M. tuberculosis,27 and there is evidence that M. tuberculosis-specific CD4+ T cells that localize in the lung parenchyma have greater capacity to control infection than CD4+ T cells present in the vasculature.28 Furthermore, the protective efficacy of mucosal-delivered BCG vaccination has been associated with the development of lung-resident CD4+ T cells,29,30 including Antigen 85B-specific CD4+ TRM.29,30 Because of their location in the mucosal tissues pathogen-specific CD4+ TRM are considered to be the first line of defense, providing a timely in situ response that can help recruit other immune cells to the site of infection.31
In this work, we demonstrated that immunization of mice with a rIAV-vectored TB vaccine led to the long-term retention of M. tuberculosis-specific tissue-resident memory CD4+ T cells in the lung. This is dependent on mucosal immunization since immunization by the i.p. route with the PR8.p25 vaccine failed to induce such responses. Further, priming by i.p. immunization with PR8.p25 followed by intranasal X31-p25 vaccine was not associated with protection (data not shown), while priming and boosting by the intranasal route with these rIAV vaccines did protect against M. tuberculosis.26 This is consistent with the fact that intranasal, but not parenteral, immunization with BCG was necessary to induce T-cell responses in the airway luminal compartment.32 Similarly, Perdomo et al.30 showed that intratracheal vaccination with BCG induced significantly higher numbers of CD4+ and CD8+ T cells to the airways than subcutaneous immunization.
The P25 CD4+ T cells present in the lung parenchyma of the mice immunized by PR8.p25 exhibited the phenotype associated with tissue-resident memory T cells, namely expression of CD69 in the absence of recent antigen stimulation. Of note, the CD4+ memory T cells induced in the lung by the PR8.p25 vaccine did not express the marker CD103, commonly co-expressed with CD69 in CD8+ TRM cells in some tissues.33 The rIAV-induced P25 CD4+ TRM cells expressed high levels of CD11a, which is also present on parenchymal CD4+ memory T cells that develop after respiratory virus34 and bacterial28 infections. The P25 CD4+ cells were scattered throughout the lung, from areas close to the pleura to those adjacent to the more central airways and blood vessels, consistent with their putative role as the first line of defense at the site of infection. Furthermore, the fact that the persistence of these cells was not affected by FTY720-induced lymphopenia is a strong indication that these are indeed long-lived tissue-resident memory cells that do not circulate through lymphoid organs and are not dependent on replenishment by circulating cells.
The CD69+ P25 cells persisting in the lungs 6 weeks after immunization with PR8.p25 showed changes in gene expression that have been identified in transcriptional profiling of CD4+ and CD8+35,36,37 TRM, including downregulation of Klf2 and S1p1r genes. Interestingly, the upstream regulator gene Foxo1 was only modestly downregulated in the lung CD69+ P25 cells, and this may be due to this gene being regulated by non-transcriptional mechanisms, such as phosphorylation and ubiquitination.38 Altogether with the high levels of expression of the pro-survival markers, Bcl-2 and Bcl-xL, these data indicate that the rIAV vaccine-induced CD69+ P25 cells can establish a long-term residency in the lungs of immunized mice.
Strikingly the enumeration of P25 T cells in digested lungs using flow cytometry was a gross underestimate (average 66 times less) of the total number of P25 T cells identified by 2-Photon microscopy. These results are in close concordance with previous report that detected 69-fold more pathogen-specific CD8+ memory T cells by quantitative immunofluorescence microscopy, compared to flow cytometry in the lungs of mice infected with LCMV.7 Considering that the loss of cells by digestion of the lungs may affect the resident cell population more than circulating cells, these results reinforce the notion that studies on tissue-resident memory cells will benefit greatly from including imaging of the cleared tissues to provide a more reliable quantification of T cells and their location. Although flow cytometry is a suitable method to evaluate differences in the frequency of cytokine-producing cells responding to antigen recall between experimental groups as done in this study, a precise quantification of the cytokine responses of the T cells identified by imaging of the lungs will require further microscopy studies using cytokine reporter tools.
Although flow cytometry may underestimate the number of parenchymal T cells, we found that P25-specific CD4+ T cells induced by the rIAV vaccine were retained in the lung parenchyma at higher numbers than those generated by pulmonary delivery of BCG.30 This may be related to the adoptive transfer strategy used in this study, and this may not exactly reflect the magnitude of the antigen-specific T-cell response generated in naive mice from the fewer endogenous p25-specific precursors. Nevertheless, we obtained similar numbers of P25 CD4+ TRM cells in the lungs 6 weeks after immunization with PR8.p25 regardless of whether the adoptive transfer was performed using 104 or 400 P25 cells (data not shown), suggesting that the generation of TRMs may be more dependent on the quality of the effector immune response generated rather than on the number of precursors present at the time of immunization. In addition, these P25 TRM CD4+ T cells were still detected 36 weeks after rIAV immunization, longer than described for other models of immunization.30
The lung CD69+ memory p25 cell subset that persisted after PR8.p25 immunization had higher levels of Tbx21 and Ifng mRNA expression than the splenic TEM cells. This elevated expression of Th1-associated genes indicates these lung-resident CD69+ memory T cells are poised for cytokine production upon recall at the mucosal sites and, therefore, likely to contribute with faster and stronger protective immune responses against future pulmonary infection. In fact, after challenge with M. tuberculosis the parenchymal P25 CD4+ TRM cells induced by immunization with rIAV vaccine produced polyfunctional cytokine responses much earlier than the P25 CD4+ T cells generated by the M. tuberculosis even when circulating CD4+ T cells were substantially depleted by FTY720 treatment. This is in concordance with previous study using FTY720 that showed that memory T cells present in the lungs following BCG immunization were sufficient to protect mice from a subsequent BCG challenge29 and with report by Sakai and colleagues showing that M. tuberculosis-specific CD4+ T cells isolated from the lung parenchyma were more protective against subsequent M. tuberculosis challenge than their counterparts from the lung vasculature when adoptively transferred into naive TCRa−/− mice.28 In our study, not only the P25 CD4+ TRMs induced by the rIAV vaccine showed early protection against challenge in the absence of circulating cells, but also were able to overcome the increased susceptibility to infection owing to the delayed CD4+ T-cell response caused by previous FTY720 treatment. Although we cannot completely exclude that part of this protection may have been due to non-antigen-specific effects of the X31-p25 rIAV, we consider this very unlikely as we (Supplementary Fig. S8) and others25 have previously shown that WT PR8 IAV does not have a protective effect against M. tuberculosis infection. Overall, these results strengthen the idea that developing vaccines with maximum potential to induce TRM populations at the sites of infection may be more effective than those inducing merely high circulating memory responses.
In a recent study assessing the ability of currently licenced influenza vaccines ability to generate TRM, intranasal administration of live attenuated influenza A virus (Flu Mist) induced virus-specific CD4+ and CD8+ TRM cells in the lungs, while the injectable, inactivated vaccine Fluzone or the live attenuated vaccine given by the intraperitoneal or subcutaneous routes induced mainly strain-specific neutralizing antibodies.39 Furthermore intranasal delivery of the inactivated vaccine failed to generate the increased CD3+CD69+ memory T cells observed in the lungs after the immunization with the live attenuated vaccine. These results indicate that the ability to provide long-term protection against vaccine and non-vaccine viral strains in mice is linked to the establishment of a stable population of long-term TRM cells by targeting the respiratory track with live attenuated viruses.
Another influenza A virus-based TB vaccine, TB/FLU-04L (Research Institute for Biological Safety Problems, Kazakhstan), has been developed, completed a phase I and is currently in a phase II clinical trial. This replication-deficient influenza vector, which expresses the M. tuberculosis-specific proteins ESAT6 and Ag85A, was well tolerated when administered by the intranasal route and induced nasal antigen-specific CD4+ and CD8+ T-cell responses.40 Here, we have shown that a rIAV TB vaccine that induces a potent immune CD4+ T-cell response to the M. tuberculosis antigen generated a long-lasting population of M. tuberculosis-specific CD4+ TRMs in the lungs of the immunized mice, which readily produce cytokines in response to M. tuberculosis challenge and are associated with significant protection in the lung. Therefore, the use of live attenuated rIAV–based TB vaccines may be an attractive vaccine strategy, either as primary pulmonary vaccine or to boost BCG in order to stimulate long-lasting TRM cells in the lungs that can respond in situ as the first line of defense against pulmonary M. tuberculosis infection.
Methods
Influenza recombinant virus
DNA encoding the M. tuberculosis Ag85B(Rv1886c)240–254 CD4+ T-cell epitope (FQDAYNAAGGHNAVF, p25) was inserted into the neuraminidase (NA) stalk of the H1N1 PR8 influenza virus between amino acids 43 and 44.20 rIAV PR8.p25 was generated using an eight-plasmid reverse genetics system, as previously described.41 Briefly, 1 µg of 8 pHW2000 DNA plasmids containing each influenza gene segment, including the pHW2000 plasmid encoding the genetically modified NA segment, were transfected into a co-culture of human embryonic kidney 293 (HEK293T) cells and Madin–Darby canine kidney cells (MDCK). Viruses were then injected into 10-day-old embryonated eggs, and after 48 h incubation, recombinant viruses were harvested from the allantoic fluid and virus titers determined using a plaque assay.42 The presence of recombinant virus was determined using a hemagglutination assay, and insertion of the appropriate epitopes was confirmed by sequencing.
Mice
Six- to eight-week-old female C57BL/6 were purchased from the Australian BioResources (NSW, Australia). P25 CD4+ TCR transgenic mice (specific for residues 240–254 of M. tuberculosis Ag85B) were kindly provided Dr. J Ernst (New York University, NY). P25 CD4+ TCR transgenic mice expressing both the M. tuberculosis p25 epitope and GFP under the control of the Ubiquitin promoter (P25-GFP) were kindly provided by Prof WR Heath (Peter Doherty Institute, University of Melbourne, Australia). The mice were maintained in specific pathogen-free conditions and bred at Centenary Institute. All mouse experiments were approved by the Sydney Local Health District Animal Ethics Committee.
Mouse immunizations, treatments, and infection
In all, 5 × 104 P25 Tg or P25-GFP cells were transferred by i.v. injection into recipient C57BL/6 mice 1 day prior to immunization. Mice were infected intranasally with 20 pfu of PR8.p25 or 104 pfu of X31.p25 and wild-type IAVs in 50 µl of saline solution. In some experiments, 104 pfu PR8.p25 were injected i.p. in a volume of 200 µl of saline solution. Intravascular staining was performed by injecting mice i.v. with 5 µg of APC-Cy7-conjugated rat anti-mouse CD45 antibody 3–5 min before being euthanized.
FTY720 (Sigma) was administered by i.p., 1 mg/kg in PBS, daily for 20 days. To assess the protective efficacy of the vaccines the mice were challenged with 100 cfu of M. tuberculosis H37Rv using a Middlebrook airborne infection apparatus20 (Glas-Col, IN, US).
Lung and spleen processing
Lungs were digested in RPMI/10%FCS containing collagenase IV at 100 U/ml and DNase I at 25 µg/ml for 30 min at 37 °C. Spleens were disrupted by passage through 70 µm cell strainers and washed with RPMI/10% fetal calf serum (FCS). Lung and spleen cell suspensions were incubated with 1 ml of ACK lysis buffer for 2 min to remove red blood cells, washed and resuspended in appropriate volume of RPMI/10%FCS.
IFNγ ELISpot
Cells were cultured at a density of less than 2 × 106 cells/ml of RPMI/10%FCS in an ELISpot plate pre-coated with 15 µg/ml of anti-mouse IFNγ monoclonal antibody (clone AN18) in the presence of pathogen-specific peptides, NP366–374 and Ag85B240–254 (Genscript), at a final concentration of 10 µg/ml or with BCG lysate at a final concentration of 5 µg/ml. As controls, cells were incubated with media alone. After 18 h of incubation plates were thoroughly washed with PBS/0.01%Tween20 and incubated with biotinylated anti-mouse IFNγ monoclonal antibody (clone XMG1.2) at final concentration 2.5 µg/ml for 2 h at 37 °C. Development was achieved by incubation with avidin-conjugated alkaline phosphatase (Sigma) followed by addition of AP conjugate substrate (Biorad). The numbers of spots were determined using an AID ELISpot Reader.
Flow cytometry
Two million cells were incubated with 1.25 µg/ml of anti-CD32/CD16 (eBiosciences) in FACS wash buffer (PBS/2%FCS/0.1%NaN3) for 30 min to block Fc receptors, washed and incubated for 30 min with a mix of fluorescent antibodies. The following antibodies were used: PerCPCy5.5-CD3 (BD Biosciences), Alexa700-CD4 (BD Biosciences), PE-CD69 (BD Biosciences), efluor450-CD62L (BD Biosciences), FITC-CD44 (Biolegend), PECy7-KLRG1 (BD Biosciences), and BV510-CD11a (BD Biosciences). Blue fixable cell death stain (Invitrogen) was added to the antibody mix at dilution recommended by manufacturer to allow dead cell discrimination. Surface markers were analyzed using the gating strategy described in Supporting Fig. S1 and FlowJo Bolean gating tool.
For intracellular staining 4 × 106 spleen cells were incubated overnight in the presence of 10 µg/ml of Ag85B240–254 peptide (Genscript), and 4 h prior to collection 10 µg/ml of BrefeldinA was added to each well. Cells were stained with anti-CD3, anti-CD4 antibodies and blue fixable cell death stain, fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences) prior to staining with anti-cytokine antibodies (PECy7-IFNγ, APC-TNF, and BV510-IL2, BD Biosciences) for 20 min. Cells were then washed with Permwash (BD Biosciences) and resuspended in FACS buffer. Cytokine expression was analyzed using the gating strategy described in Supporting Fig. S2 and FlowJo Bolean gating tool.
Samples were run on a LSR Fortessa Flow cytometer. Analysis was performed using FlowJo software (TreeStar, Ashland, OR)
Lung clarification and two photon microscopy
Lungs were fixed with 10% formalin and cleared for at least 12 h with RapiClear1.52 (Sunjilab, Taiwan). The cleared lungs were mounted into slides fitted with iSpacers (Sunjilab, Taiwan) filled with the clearing reagent before analysis using a Nikon Ti-U inverted fluorescence microscope with a 25x Nikon water immersion objective (1.1 NA, WD 2 mm) and a dedicated single-beam LaVision TriM Scope I (LaVision Biotec: Germany) controlled by Imspector software (LaVision). The microscope was outfitted with one Ti:Sapphire laser (MaiTai DeepSee: Spectra-physics) and one Synchronously Pumped Optical Parametric Oscillator (OPO) pumped by a Chameleon Ultra II Ti:Sapphire laser (Coherent). Emission wavelengths were collected with ultrasensitive photomultiplier tubes (GaSP Hamamatsu) for 412–472 nm (second harmonic signal), 490–520 nm (GFP), 573–633 nm (A594). The DeepSee laser was tuned to 940 nm and OPO was tuned to 1080 nm. Parfocal alignment of lasers was adjusted using fluorescent beads before each experimental session. Data correction and analysis were conducted using ImarisJ software (Bitplane, South Windsor, CT). For detection of vascular vessels mice were injected i.v. with Alexa 594-conjugated wheat germ agglutinin 30 min prior to organ collection. For quantification of GFP-positive cells in the lungs, 2 µm step z-stacks of 400 x 400 x 1200 µm random areas of the right lobes were acquired. Quantification was done using the Imaris11 spot tool based on at least seven random z-stacks from each lung sample.
Cell isolation and transcriptional profile analysis
Different P25 T-cell subsets were sorted in a BD FACSAria II into: lung-resident memory (L-RM, CD69+), spleen effector memory (S-EM, CD69-CD62L-) from a pool of cells obtained from immunized mice (n = 20) 6 weeks after immunization. Effector P25 cells (S-eff) were obtained from mice that have been infected with PR8.p25 for 11 days. Following RNA extraction (QIAGEN RNeasy Mini), and DNase I (New England Biolabs) digestion, cDNA were prepared using Tetro cDNA synthesis kit (Bioline). Quantitative real-time PCR of Klf2, S1pr1, Foxo1, Tbx21, Bcl-2, Bcl-xL, and Ifng was performed with SensiFAST SYBR Hi-ROX mix and the following primers: 18 s Fw GTAACCCGTTGAACCCCATT, Rv CCATCCAATCGGTAGTAGCG; Klf2 Fw-CTAAAGGCGCATCTGCGTA, Rv TAGTGGCGGGTAAGCTCGT; S1pr1 Fw CACAGGCAAGTTGAACATCG, Rv GGATGATGAAGCAGCAGATG; Foxo1 Fw TGTCAGGCTAAGAGTTAGTGAGCA, Rv GGGTGAAGGGCATCTTTG; Tbx21 Fw GCCAGGGAACCGCTTATATG, Rv GACGATCATCTGGGTCACATT; Bcl-2 Fw CCGGGAGAACAGGGTATGATAA, Rv CCCACTCGTAGCCCCTCTG; Bcl-xL Fw CACTGTGCGTGGAAAGCGTA, Rv AAAGTGTCCCAGCCGCC; IFNg Fw ACAATGAACGCTACACACTGCAT, Rv TGGCAGTAACAGCCAGAAACA. The threshold cycle of individual genes was normalized to the value of 18s rRNA (ΔCT) for each cell population and gene expression calculated by the 2(−ΔCT) method.
Bacterial quantification
Bacterial quantification was performed by plating serial dilutions of lung homogenates onto Middlebrook 7H11 supplemented with 10% oleic acid-albumin enrichment (Difco) agar plates and culture for 3 weeks at 37 °C.
Statistical analysis
Data were analyzed using Student’s t-test when comparing two experimental groups or ANOVA followed by Tukey post-test correction for more than two groups. Differences with a value of p < 0.05 were considered statistically significant.
References
World Health Organization, Geneva, Switzerland. Global Tuberculosis Report. (2017).
Fine, P. E. BCG: the challenge continues. Scand. J. Infect. Dis. 33, 243–245 (2001).
Reiley, W. W. et al. Distinct functions of antigen-specific CD4 T cells during murine Mycobacterium tuberculosis infection. Proc. Natl Acad. Sci. U.S.A. 107, 19408–19413 (2010).
Pawlowski, A., Jansson, M., Skold, M., Rottenberg, M. E. & Kallenius, G. Tuberculosis and HIV co-infection. PLoS Pathog. 8, e1002464 (2012).
Tameris, M. D. et al. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet https://doi.org/10.1016/S0140-6736(13)60177-4 (2013).
Masopust, D., Vezys, V., Marzo, A. L. & Lefrancois, L. Preferential localization of effector memory cells in nonlymphoid tissue. Science 291, 2413–2417 (2001).
Steinert, E. M. et al. Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell 161, 737–749 (2015).
Schenkel, J. M. & Masopust, D. Tissue-resident memory T cells. Immunity 41, 886–897 (2014).
Allende, M. L., Dreier, J. L., Mandala, S. & Proia, R. L. Expression of the sphingosine 1-phosphate receptor, S1P1, on T-cells controls thymic emigration. J. Biol. Chem. 279, 15396–15401 (2004).
Carlson, C. M. et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature 442, 299–302 (2006).
Mackay, L. K. et al. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J. Immunol. 194, 2059–2063 (2015).
Wu, T. et al. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 95, 215–224 (2014).
Gebhardt, T. et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10, 524–530 (2009).
Thom, J. T., Weber, T. C., Walton, S. M., Torti, N. & Oxenius, A. The salivary gland acts as a sink for tissue-resident memory CD8(+) T cells, facilitating protection from local cytomegalovirus infection. Cell Rep. 13, 1125–1136 (2015).
Sheridan, B. S. et al. Oral infection drives a distinct population of intestinal resident memory CD8(+) T cells with enhanced protective function. Immunity 40, 747–757 (2014).
Fernandez-Ruiz, D. et al. Liver-resident memory CD8+T cells form a front-line defense against malaria liver-stage infection. Immunity 45, 889–902 (2016).
Turner, D. L. et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. . Mucosal Immunol. 7, 501–510 (2014).
Strutt, T. M. et al. IL-15 supports the generation of protective lung-resident memory CD4 T cells. Mucosal Immunol. https://doi.org/10.1038/mi.2017.101 (2017).
Oja, A. E. et al. Trigger-happy resident memory CD4(+) T cells inhabit the human lungs. Mucosal Immunol. https://doi.org/10.1038/mi.2017.94 (2017).
Florido, M. et al. Epitope-specific CD4+, but not CD8+, T-cell responses induced by recombinant influenza A viruses protect against Mycobacterium tuberculosis infection. Eur. J. Immunol. 45, 780–793 (2015).
Anderson, K. G. et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9, 209–222 (2014).
Kerdiles, Y. M. et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat. Immunol. 10, 176–184 (2009).
Chiba, K. et al. Fingolimod (FTY720), sphingosine 1-phosphate receptor modulator, shows superior efficacy as compared with interferon-beta in mouse experimental autoimmune encephalomyelitis. Int. Immunopharmacol. 11, 366–372 (2011).
Morris, M. A. et al. Transient T cell accumulation in lymph nodes and sustained lymphopenia in mice treated with FTY720. Eur. J. Immunol. 35, 3570–3580 (2005).
Redford, P. S. et al. Influenza A virus impairs control of Mycobacterium tuberculosis coinfection through a type I interferon receptor-dependent pathway. J. Infect. Dis. 209, 270–274 (2014).
Muflihah, H. et al. Sequential pulmonary immunization with heterologous recombinant influenza A virus tuberculosis vaccines protects against murine M. tuberculosis infection. Vaccine 36, 2462–2470 (2018).
Srivastava, S. & Ernst, J. D. Cutting edge: Direct recognition of infected cells by CD4 T cells is required for control of intracellular Mycobacterium tuberculosis in vivo. J. Immunol. 191, 1016–1020 (2013).
Sakai, S. et al. Cutting edge: control of Mycobacterium tuberculosis infection by a subset of lung parenchyma-homing CD4 T cells. J. Immunol. 192, 2965–2969 (2014).
Connor, L. M. et al. A key role for lung-resident memory lymphocytes in protective immune responses after BCG vaccination. Eur. J. Immunol. 40, 2482–2492 (2010).
Perdomo, C. et al. Mucosal BCG vaccination induces protective lung-resident memory T cell populations against tuberculosis. MBio 7, https://doi.org/10.1128/mBio.01686-16 (2016).
Turner, D. L. & Farber, D. L. Mucosal resident memory CD4 T cells in protection and immunopathology. Front. Immunol. 5, 331 (2014).
Horvath, C. N., Shaler, C. R., Jeyanathan, M., Zganiacz, A. & Xing, Z. Mechanisms of delayed anti-tuberculosis protection in the lung of parenteral BCG-vaccinated hosts: a critical role of airway luminal T cells. Mucosal Immunol. 5, 420–431 (2012).
Casey, K. A. et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188, 4866–4875 (2012).
Teijaro, J. R. et al. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187, 5510–5514 (2011).
Mackay, L. K. et al. The developmental pathway for CD103(+)CD8+tissue-resident memory T cells of skin. Nat. Immunol. 14, 1294–1301 (2013).
Skon, C. N. et al. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+T cells. Nat. Immunol. 14, 1285–1293 (2013).
Kumar, B. V. et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20, 2921–2934 (2017).
Huang, H. & Tindall, D. J. Regulation of FOXO protein stability via ubiquitination and proteasome degradation. Biochim. Biophys. Acta 1813, 1961–1964 (2011).
Zens, K. D., Chen, J. K. & Farber, D. L. Vaccine-generated lung tissue-resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight 1, https://doi.org/10.1172/jci.insight.85832 (2016).
Walker, K. B. et al. Novel approaches to preclinical research and TB vaccine development. Tuberc. (Edinb.) 99(Suppl 1), S12–S15 (2016).
Hoffmann, E., Krauss, S., Perez, D., Webby, R. & Webster, R. G. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20, 3165–3170 (2002).
Cukalac, T. et al. Narrowed TCR diversity for immunised mice challenged with recombinant influenza A-HIV Env(311-320) virus. Vaccine 27, 6755–6761 (2009).
Acknowledgements
This work was supported by the National and Medical Research Council of Australia through Project Grant APP1044343, and the Center of Research Excellence in TB Control (APP1043225), and the NSW Government through its infrastructure grant to the Centenary Institute. H.M. was a recipient of Australia Award Scholarship from the Australian Department of Foreign Affairs and Trade.
Author information
Authors and Affiliations
Contributions
M.F., H.M. and L.L designed and performed experiments, and M.F, wrote the manuscript. Y.X. developed the rIAV vaccines under supervision of J.S. F.S. assisted with 2-photon microscopy experiments. M.P., C.F., P.B. and J.A.T. contributed to the experimental design and provided intellectual input. W.B. was responsible for study design, data interpretation, and study supervision. All the authors contributed to the editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Flórido, M., Muflihah, H., Lin, L.C.W. et al. Pulmonary immunization with a recombinant influenza A virus vaccine induces lung-resident CD4+ memory T cells that are associated with protection against tuberculosis. Mucosal Immunol 11, 1743–1752 (2018). https://doi.org/10.1038/s41385-018-0065-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-018-0065-9
This article is cited by
-
BCG-booster vaccination with HSP90-ESAT-6-HspX-RipA multivalent subunit vaccine confers durable protection against hypervirulent Mtb in mice
npj Vaccines (2024)
-
Tissue resident memory T cells in the respiratory tract
Mucosal Immunology (2022)
-
Mucosal immunization with a delta-inulin adjuvanted recombinant spike vaccine elicits lung-resident immune memory and protects mice against SARS-CoV-2
Mucosal Immunology (2022)
-
Intrapulmonary vaccination with delta-inulin adjuvant stimulates non-polarised chemotactic signalling and diverse cellular interaction
Mucosal Immunology (2021)
-
Intratracheal inoculation of AHc vaccine induces protection against aerosolized botulinum neurotoxin A challenge in mice
npj Vaccines (2021)