Abstract
The tissue dendritic cell (DC) compartment is heterogeneous, and the ontogeny and functional specialization of human tissue conventional DC (cDC) subsets and their relationship with monocytes is unresolved. Here we identify monocyte-related CSF1R+Flt3- antigen presenting cells (APCs) that constitute about half of the cells classically defined as SIRPα+ DCs in the steady-state human small intestine. CSF1R+Flt3- APCs express calprotectin and very low levels of CD14, are transcriptionally related to monocyte-derived cells, and accumulate during inflammation. CSF1R+Flt3- APCs show typical macrophage characteristics functionally distinct from their Flt3+ cDC counterparts: under steady-state conditions they excel at antigen uptake, have a lower migratory potential, and are inefficient activators of naïve T cells. These results have important implications for the understanding of the ontogenetic and functional heterogeneity within human tissue DCs and their relation to the monocyte lineage.
Similar content being viewed by others
Introduction
Dendritic cells (DCs) shape the adaptive immune system in response to environmental cues and play a crucial role in balancing tolerance and inflammatory immune responses. Based on ontogeny, DCs divide into plasmacytoid (p) DCs producing type I interferons in response to viral stimuli, and conventional (c) DCs that comprise a developmentally and functionally heterogeneous population of cells that by definition capture and process antigens, migrate to lymph nodes and stimulate T cell responses through antigen presentation.1,2
According to recent studies in both mice and humans a macrophage/dendritic progenitor gives rise to both M-CSF-dependent monocytes and common dendritic progenitors (CDP) in the bone marrow.3,4 CDPs develop into both pre-pDCs and pre-conventional DCs (cDCs) in a Flt3 ligand (Flt3L)-dependent manner.4,5,6 Pre-cDCs enter the circulation committed to differentiate into one of the two major cDC subsets.7,8,9 In human blood these subsets are distinguished by their selective expression of CD141 (cDC1) or CD1c (cDC2).10 The development of cDC1 is dependent on IRF8, ID2 and BATF3,11,12 while cDC2 develop in an IRF4-dependent manner.13,14 Both pre-DCs and DCs have a very short half live in the circulation, suggesting that they rapidly emigrate to peripheral tissues after generation.15
The exposure to diverse dietary and microbial antigens constantly challenges the local immune system in the gastrointestinal tract, and intestinal DCs play an essential role in maintaining homeostasis. In mice, intestinal DCs are subdivided based on expression of CD11b and CD103, and humanized mouse models and transcriptomic analyses indicate that SIRPα (CD172a) and CD103 define corresponding subsets in the human small intestine (hSI).13,16,17 hSI SIRPα-CD103+ DCs resemble murine intestinal BATF3-dependent CD11b-CD103+ DCs as well as CD141+ human blood DCs, and can thus be assigned to the cDC1 lineage.16,18 They express Clec9a, CD141, XCR1, and TLR3, which are considered conserved markers specific for cDC1 across a variety of tissues and species, but do not express CD11b or SIRPα.19,20,21 In contrast, hSI SIRPα+CD103+ DCs expressing CD11b and CD1c are IRF4-dependent,13,22 and belong to the cDC2 lineage.16,17
A third DC subset described in the hSI, SIRPα+CD103- DCs, transcriptionally resembles human and murine monocytes, and, to a lesser extent, murine intestinal CD11b+CD103- DCs.16 Murine tissue CD11b+CD103- DCs are partly M-CSF-dependent, express CCR2, CX3CR1, CD64, and F4/80, and can exhibit Mf-like tissue residency and slow turnover.22,23,24,25 However, another subset of murine CD11b+CD103- DCs is Flt3L-dependent, lacks F4/80, and migrates to mesenteric lymph nodes (MLNs).22,26 These findings indicate that the murine subset of CD11b+CD103- consists of both monocyte-derived cells and pre-cDC-derived cells. Studies in mice suggest that monocytes not only give rise to macrophages (Mfs), but also differentiate into DCs in various tissues including intestinal mucosa.24,27 Initial attempts to corroborate these findings in humans have identified a subset of CCR2+SIRPα+CD103- DCs,26 but the ontogeny and characteristics of this subset are unclear, and whether monocytes contribute to the intestinal DC compartment remains unresolved.
By applying extensive phenotyping and transcriptional profiling we find that a substantial fraction of antigen presenting cells (APCs) within the typical SIRPα+ DC compartment in the hSI do not express Flt3 and are likely derived from blood monocytes. Despite phenotypic similarity, intestinal Flt3- APCs in the CD14-CD11c+SIRPα+ population exhibit a higher capacity for antigen processing, and inferior potential for migration and priming of naïve T cells than their Flt3+ counterparts in hSI.
Results
Differential expression of SIRPα and CD103 defines distinct hSI DC populations
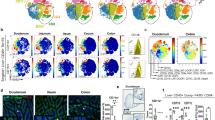
Human tissue DCs have previously typically been defined as CD11c+CD14- cells among the CD45+HLA-DRhi APCs, based on morphology, phenotype, and functional properties.16,28,29,30 Therefore, we adopted this widely used strategy and gated on live CD45+HLA-DR+CD11c+CD14- cells and separated subsets of DCs using SIRPα and CD103 (Fig. 1a, upper panel) as previously described.16 We verified that among CD45+HLA-DR+CD11c+ cells in blood, CD14hi cells represent CSF1R+Flt3- monocytes, while CD14- cells represent CSF1R-Flt3+ cDCs (Supplementary Figure 1A), and used this gating strategy for phenotypic comparison with hSI subsets (Fig. 1a, lower panel).
Differential expression of SIRPα and CD103 defines distinct hSI DC populations. a Lamina propria cells and PBMCs were gated on live CD45+HLA-DR+ cells, and the nominal DC compartment was identified as CD14-CD11c+. DC subsets were defined by SIRPα and CD103 expression. b IRF8 and IRF4 expression throughout hSI DC subsets. Vertical lines represent gates set using isotype-matched control antibody. Data are representative of at least three donors. c Relative abundance of hSI DC subsets among live CD45+HLA-DRhiCD14-CD11c+ cells (N = 72). d Relative abundance of DC subsets, stratified according to donor SI histopathology (N = 65). * P < 0.05 *** P < 0.001 (RM-ANOVA). e Expression levels of Flt3 and CSF1R within the main hSI APC subsets as gated in the plot to the right. Data are representative of at least three subjects. See Supplementary Figure 4D for full gating strategy of APC subsets
hSI SIRPα+CD103+ DCs expressed IRF4 and CD1c, similar to blood cDC2, whereas the SIRPα+CD103- subset expressed both markers at lower levels (Fig. 1b and Supplementary Figure 1B). Within the SIRPα- subsets, the CD103+ DCs expressed IRF8, CD141+ and Clec9a+, comparable to blood cDC1, while the CD103- cells contained IRF8-, CD141-, and Clec9a- cells that represented contaminating CD19+ B cells (Fig. 1b and Supplementary Figure 1B-C), while T cells were not present within the CD45+HLA-DR+CD11c+CD14- DC gate (data not shown). SIRPα-CD103-Clec9a+ cells constituted a very small fraction of DCs and were not studied further.
SIRPα+CD103+ DCs were most abundant (Fig. 1c), and the relative representation of the subsets was independent of age or gender (data not shown). Following evaluation by an experienced pathologist, the majority of the 105 samples were graded as non-inflamed, whereas nine cases were graded as unspecific low-grade inflammation characterized by infiltration of neutrophils and increased numbers of mononuclear cells. Interestingly, in the latter samples SIRPα+CD103- DCs were significantly enriched (Fig. 1d). Of note, blood monocytes as well as hSI CD14+CD11c+ (recently recruited) and CD14+CD11c- (mature) tissue Mfs also express SIRPα.31 As monocytes are recruited to inflamed tissues we hypothesized that SIRPα+CD103-CD14- APCs contain monocyte-derived cells, and therefore analyzed the expression pattern of typical monocyte-associated, DC-associated, and Mf-associated surface markers across the DC subsets.
Pre-cDC-derived cells depend on Flt3L for development, contrasting the M-CSF-dependent development of monocyte-derived cells.32,33 All DC subsets expressed variable levels of the Flt3L-receptor Flt3, while CD14+CD11c+ Mf and CD14+CD11c- Mf were Flt3- (Fig. 1e, upper panel). None of the DC subsets expressed the classical Mf markers CD163 and MerTK (Supplementary Figure 2A). In addition to CD14+CD11c+ Mf and a fraction of CD14+CD11c- Mf, also SIRPα+ DCs expressed CSF1R (Fig. 1e, lower panel). As the detected CSF1R levels were generally low, we aimed to establish an alternative reliable marker for monocyte-derived cells. CD64, CCR2, and CX3CR1 are described to identify monocyte-derived tissue cells in mice.28,34,35,36 These surface receptors were expressed by a subpopulation of both SIRPα+ DC subsets, while SIRPα- tissue cDC1 were negative (Supplementary Figure 2A). However, they proved not to be specific for monocyte-derived cells in humans as also subsets of blood cDC2 expressed CD64, CCR2, and CX3CR1 (Supplementary Figure 2B).
Calprotectin expression identifies Flt3- cells within the SIRPa+ DC compartment
We have previously shown that calprotectin (S100A8/A9) is expressed by monocytes and a subset of gut Mf, and that levels of calprotectin expression negatively correlated with Mf differentiation.31 This prompted us to assess the usefulness of calprotectin as marker for recently monocyte-derived tissue cells in the current context. We analyzed its expression across all cells in hSI (Supplementary Figure 3A) and peripheral blood (Supplementary Figure 3B). Only two distinct populations of calprotectin+ cells were present in both compartments: CD45intHLA-DR-CD15+ cells, as well as CD45highHLA-DR+CD11c+CD14low-hi APCs consistently comprised ~95% of all calprotectin+ cells detected in the hSI (Supplementary Figure 3A).
More specifically, among hSI APCs calprotectin was expressed by CD14+CD11c+ Mf, a small subset of CD14+CD11c-/int Mf, and a subpopulation of both CD14- SIRPα+ subsets, but not by SIRPα- DCs (Fig. 2a). SIRPα+CD103- APCs consistently contained higher numbers of calprotectin+ cells than SIRPα+CD103+ APCs, and the expression levels of calprotectin correlated inversely with CD103 expression within the SIRPα+CD103+ population (Supplementary Figure 3C). Expression levels of CD45, HLA-DR, and CD11c, as well as of the cDC2-associated surface markers SIRPα and CD1c, and the occasionally described FcɛRI and CD2, were overlapping between calprotectin- and calprotectin+ SIRPα+ APCs in hSI (Supplementary Figure 3D). Of note, in contrast to calprotectin-SIRPα+ DCs, calprotectin+SIRPα+ APCs exhibited very low levels of Flt3 (Fig. 2b). Costaining of calprotectin with CSF1R was not possible as the CSF1R signal was lost when attempting to combine with intracellular staining (data not shown). The reciprocal relationship between calprotectin and Flt3 was affirmed by RNA sequencing of the sorted subsets (see Fig. 5d below). Moreover, analysis of available transcriptomics data of DC subsets sorted from hSI16 revealed a similar relationship between calprotectin and Flt3 (Supplementary Figure 3E). These findings suggest that CD14-SIRPa+ calprotectin+ APCs are contained within the typical cDC2 “gate,” but that they do not represent pre-cDC-derived Flt3+ DCs.
Calprotectin expression defines a subset of Flt3- cells. a Abundance of calprotectin+ cells among the main subsets of CD14-CD11c+ DCs, CD14+CD11-, and CD14+CD11c+ Mf (N = 17). b Expression of Flt3 in DC compartment subdivided by SIRPα and calprotectin expression; gates were set based on isotype-matched control antibody (calprotectin) or FMO (SIRPα) (N = 4). * P < 0.05 ** P < 0.01 (RM-ANOVA). c Abundance of calprotectin+ cells among DC subsets, stratified according to SI histopathology (N = 9). ** P < 0.01 *** P < 0.001 (t-test). d Calprotectin expression by blood monocytes and DC subsets (representative of six subjects), and (e) blood-derived cDC2 and cocultured allogeneic lamina propria cells (LPLs) after culture with/without addition of intestinal epithelial cell supernatant (IECsup) for 24 h (representative of three independent experiments)
We found that on average 40% of cells within the SIRPα+ DC compartment were calprotectin+, and enriched in, but not confined to the CD103- subset. In accordance with earlier reports that calprotectin+ monocyte-derived cells were found in increased numbers in acute and chronic intestinal inflammation,37,38 the relative abundance of calprotectin+ cells among the SIRPα+ subsets was also significantly increased in tissues with histological signs of low-grade inflammation (Fig. 2c).
In blood, only neutrophils (CD15+CD16+), eosinophils (CD15+CD16-), and CSF1R+ monocytes, but not Flt3+ cDCs, expressed calprotectin (Fig. 2d and Supplementary Figure 3B). Moreover, SIRPα+ DCs isolated from blood did not upregulate calprotectin when cultured together with hSI lamina propria (LP) cells, epithelial cell derived factors, or a combination of these (Fig. 2e). In accordance with previously published work,39 these findings indicated that DCs are unlikely to upregulate calprotectin expression upon encountering the tissue environment, and suggest that the observed calprotectin+ cells among the CD45highHLA-DR+CD11c+CD14- tissue APCs, commonly termed DC, are related to infiltrating calprotectin+ monocytes.
Intestinal APCs within the DC compartment are short-lived and lose calprotectin expression in vivo
Having determined that calprotectin marks a substantial fraction of cells within the Flt3-CSF1R+SIRPα+ DC compartment, we next assessed the in vivo replacement kinetics of DCs in hSI to obtain insight in the regulation of calprotectin in tissue APCs over time. To this end we obtained mucosal biopsies from the duodenal segment of whole organ pancreas grafts 3 and 6 weeks after transplantation of type I diabetic patients.40 Chimerism of APC subsets from patients without histological signs of rejection was determined by flow cytometry using allotype-specific HLA class I antibodies (Fig. 3a). In contrast to CD14+CD11c- Mf, the compartment of CD14+CD11c+ Mf cells represents short-lived intermediaries,31,41 and was mostly of recipient origin within 3 weeks after surgery (Fig. 3a, b), confirming previous findings for hSI31 or skin early-Mf.42 DCs were almost all of recipient origin 6 weeks post-transplantation (Fig. 3a,b). Among the DC subsets, SIRPα+CD103- DCs exhibited the highest replacement rate, significantly higher than that of SIRPα-CD103+ DCs (Fig. 3a, b). We consistently found more SIRPα- DCs expressing the proliferation marker Ki67 in healthy hSI (Fig. 3c). This suggests that DCs are able to proliferate in situ in hSI, but that they, irrespective of origin, do not exhibit considerable longevity or potential for self-renewal, as virtually all hSI DCs were replaced by recipient cells 6 weeks after transplantation (Fig. 3a, b).
Replacement kinetics, calprotectin expression and proliferation of DC subsets in vivo. a Flow cytometry plots depicting donor-derived vs. recipient-derived cells among the main hSI DC subsets, CD14+CD11c+ Mf and stromal cells in the graft tissue at 6 weeks post-transplantation, representative of five subjects. b Distribution of recipient-derived cells among cell populations in the graft at 3 (N = 7) and 6 weeks (N = 5) post-transplantation. * P < 0.05 ** P < 0.01 (RM-ANOVA). c Expression of Ki67 within the main DC subsets (N = 6) in steady-state hSI (ANOVA). d Calprotectin expression in donor-derived vs. recipient-derived cells in the graft 3 weeks after transplantation (N = 3). ** P < 0.01 (RM-two-way-ANOVA). e Co-expression of calprotectin and Ki67 (red) within SIRPα+ DCs in steady-state hSI (N = 5). Gates were set based on isotype-matched control antibodies (dark-grey contour represents isotype control for Ki67). * P < 0.05 (t-test)
Importantly, while newly recruited recipient-derived SIRPα+ APCs in the graft showed calprotectin expression levels comparable to those observed in steady-state hSI, the few residual donor-derived APCs were almost completely calprotectin-negative 3 weeks post-transplantation (Fig. 3d). These data suggest that similar to other tissues,39,43 monocytes recruited to the hSI lose calprotectin expression over time. Furthermore, among the SIRPα+ DCs, calprotectin+ cells contained fewer proliferating (Ki67+) cells than calprotectin- cells (Fig. 3e).
Low-level CD14 expression distinguishes calprotectin+Flt3- APCs from calprotectin-Flt3+ DCs
Even though all cells within the APC compartment usually defined as DCs appeared to be negative for CD14 by flow cytometric analysis regardless of calprotectin expression (Fig. 1a and Supplementary Figure 3D), all calprotectin+ cells, except HLA-DR-CD15+ leukocytes, were depleted with CD14-specific magnetic bead separation of intestinal cell suspensions (Supplementary Figure 3A). This suggested low-level CD14 expression on calprotectin+ APCs in the DC compartment, and prompted us to investigate the expression of CD14 in more detail. We were able to divide the DC compartment into two populations when applying CD14-targeting magnetic beads: while virtually all SIRPα- DCs were contained in the CD14- fraction (Fig. 4a, left), SIRPα+ cells were highly enriched in the positively selected fraction within the DC gate that appeared CD14- with flow cytometry (termed CD14+/low). Calprotectin+ APCs were highly enriched in the CD14+/low DC fractions among both CD103+ and CD103- cells (Fig. 4a, right). This was irrespective of antibody clones or type of beads used for magnetic sorting (Fig. 4b), and likely due to highly increased avidity of the antibody-armed beads in combination with the strong magnetic field, as compared to divalent mIgG antibodies used for flow cytometry staining. Analysis of CD14 mRNA expression levels in bead-separated and FACS-sorted DC subsets and CD14+CD11c+ Mf confirmed that SIRPα+ DCs isolated as CD14+ by beads, but appearing in the CD14- gate by flow cytometry, indeed showed a higher CD14 mRNA expression compared to SIRPα+ DCs that were CD14- according to both flow and bead separation (Fig. 4c).
Low levels of CD14 distinguish the subset of calprotectin+Flt3- APCs within the nominal DC compartment. a Separation of hSI cell populations using CD14-specific beads, and abundance of calprotectin+ cells in the resulting live CD45+HLA-DR+ subpopulations defined by CD14, CD11c, SIRPα, and CD103 expression. Data are representative of 12 subjects. ** P < 0.01 *** P < 0.001 **** P < 0.0001 (RM-ANOVA). b Abundance of calprotectin+ cells among DC subsets after bead-based separation using different CD14 antibody clones (TUK4, 18D11; both mIgG1) or isotype control, and different bead systems (MACS or Dynabeads targeting mIgG). * P < 0.05 (RM-ANOVA). c CD14 mRNA levels in FACS-sorted tissue DC subsets and CD14+CD11c+ Mf assessed by qPCR. Tissue cells were pre-sorted using CD14-beads. DCs were sorted in both fractions as CD45+HLA-DR+CD14-CD11c+ with differential SIRPα and CD103 expression, and Mf were sorted from the CD14+ fraction as CD45+HLA-DR+CD14hiCD11c+. * P < 0.05, *** P < 0.001, **** P < 0.0001 (ANOVA). d Morphology of cell subsets sorted as described in (c) and analyzed by microscopy after cytospin and hemacolor staining. Scale bar represents 10 μm. Representative images for three subjects. e Phenotype-tSNE performed on live CD45+HLA-DR+ APCs concatenated from CD14-bead-separated fractions, with FSC, SSC, autofluorescence, CD45, HLA-DR, CD11c, CD14, SIRPα, CD103, calprotectin, and CD14-bead-reactivity as parameters (representative of three subjects). tSNE maps were either color-coded for the fluorescent intensity values of antigens (CD11c, calprotectin, CD14), or overlaid with all APC subsets (bottom right) as defined in Supplementary Figure 4E. f tSNE map zoomed in on populations of interest, with adjunct histograms for tSNE-y and tSNE-x for CD103+/- subsets
Of note, further phenotyping of the bead-separated subsets revealed that SIRPα+ cells in the CD14- fraction showed significantly higher levels of Flt3 (Supplementary Figure 4A) and lower levels of CSF1R compared to bead-isolated CD14+/lowSIRPα+ cells (Supplementary Figures 4B,C), regardless of CD103 expression. Also, the morphology of bead-separated and FACS-sorted CD14+/lowSIRPα+ DCs was more similar to CD11c+CD14+ Mf, partly with lobulated nuclei and lacking the typical dendrites displayed by CD14-SIRPα+ DCs (Fig. 4d). Both these findings indicated that CD14+/lowSIRPα+ cells although mimicking a typical cDC2 phenotype are more related to the monocyte lineage than to cDC2.
To be able to appreciate the phenotypic heterogeneity of the CD45+HLA-DR+ APC compartment, we applied an unsupervised dimensionality reduction analysis (tSNE) on single cell level with phenotyping parameters (FSC, SSC, autofluorescence, CD45, HLA-DR, CD11c, CD14, SIRPα, CD103, calprotectin, CD14-bead-reactivity) (Fig. 4e). CD11c expression determined clustering into two main subsets (Fig. 4e, top left). While a CD14 expression continuum spanned the overall APC population (Fig. 4e, bottom left), calprotectin showed an expression continuum most pronounced within the CD11c+ APCs (Fig. 4e, top right). We have recently shown that gut Mf can be divided into four distinct subsets: CD14+CD11c+ recently recruited short-lived intermediaries (Mf1 and Mf2) and mature CD14+CD11c- Mfs (Mf3 and Mf4).31 Overlaying the tSNE maps with these Mf and DC subsets (see Supplementary Figure 4D for complete gating strategy), we found that the CD14+/low APCs in the classical DC gate clustered between Mf1 and CD14- DC. CD103- and CD103+ subpopulations were largely overlapping within the CD14+/low APCs and CD14- DC clusters (Fig. 4f). Slight separation on tSNE-y was likely solely dependent on CD103, as the expression of all other markers was very similar (data not shown; the annotated dataset is publicly available for more insights from FlowRepository.org).
The overall phenotype-tSNE pattern suggests a divergence of Mf1 with high calprotectin levels (light green), branching out into either Mf2 with lower calprotectin levels (dark green) and then Mf3 (lavender), or into CD14+/lowcalprotectin+/- cells mimicking SIRPα+ DC phenotype (black).
CD14+/low APCs are enriched for monocyte gene signatures
To get further insight into the lineage relationships of bona fide DCs and potentially monocyte-derived cells within the DC gate, we performed RNA sequencing on sorted tissue cells from non-inflamed hSI specimens (N = 2). As calprotectin is an intracellular protein and therefore not useful as gating marker for subsequent RNA analysis, we applied the CD14-bead-based separation technique described above in combination with FACS to sort six distinct subsets: from the CD14+-bead-separated fraction we sorted CD14+CD11c+ Mf, CD14+/lowSIRPα+CD103+ cells, CD14+/lowSIRPα+CD103- cells; and from the CD14- bead-separated fraction we sorted CD14-SIRPα+CD103+ DCs, CD14-SIRPα+CD103- DCs and CD14-SIRPα-CD103+ DCs.
We decreased the dimensional complexity of the RNA-seq data by performing principal component analysis, revealing that all CD14+ subsets clustered and separated well from the CD14- subsets (PC1, explaining 50% of the variance; Fig. 5a). Consistent with this finding, Pearson correlation analysis together with hierarchical clustering of the differentially expressed genes showed that all CD14+ and CD14- subsets, respectively, formed separate clusters with high intracluster-correlation (Supplementary Figure 5). To further substantiate the differences between the CD14+ and the CD14- subsets, we performed 4k-means clustering analysis, which comprised more than 4500 genes (Fig. 5b). Cluster 1 comprised genes more highly expressed in the CD14+ compartment. This cluster contained monocyte-related genes such as CD14, S100A12, S100A8, S100A9, and TREM1, whereas cluster 2 and 3 contained genes more highly expressed in CD14- cells, encompassing DC-related genes such as FLT3 and ZBTB46. Clusters 1-3 therefore separate monocyte-related cells from bona fide DCs. Cluster 4 comprised genes that were downregulated within the CD14-SIRPα- DC subset. This included both monocyte and cDC2-related genes such as CD207, CD209, SIRPB, and CD1C.
Flt3- APCs are enriched for monocyte gene signatures according to RNA-seq data. a Principal component analysis on RNA-seq data generated from six subsets of hSI cells of the classical DC phenotpye and CD14+CD11c+ Mf, sorted as in Fig. 4c (N = 2). b K-means (k = 4) clustering of RNA-seq data for the cell subsets. c Overlap of clusters 1 and 2/3 from k-means clustering of RNA-seq data with core gene signatures generated from published data for human blood monocyte and cDC2 populations, respectively. d Gene expression values of calprotectin subunits S100A8, S100A9, and CD5 plotted vs. gene expression values of FLT3 generated from RNA-seq. # indicates two visually overlapping data points
In order to validate our data, we next compared the gene clusters 1–3 to published data for blood monocyte and DC populations. To minimize potential bias, we generated core gene signatures for human blood monocytes and cDCs from three different research groups as depicted in Supplementary Figures 6A and 6B.19,44,45 We defined core gene signatures as the overlap of the gene signatures derived from the individual datasets, consisting of 29 genes for CD14hi monocytes (Supplementary Table 1), and 55 genes for cDC2s (Supplementary Table 2), and validated the protein expression pattern for representative signature genes in our gating strategy for blood subsets (Supplementary Figures 6C and 6D). Importantly, cluster 1 contained 14 out of 29 genes in the blood monocyte core gene signature (hypergeometric test P value = 1.689378e-15), while clusters 2/3 covered 28 out of 55 genes (hypergeometric test P value = 2.078949e-25) in the cDC2-gene signature (Fig. 5c, Supplementary Tables S1 and S2). Only one gene in the cDC2 signature was found in cluster 1, and only one gene in the monocyte signature was detected in clusters 2/3 (Supplementary Tables S1 and S2). In addition, Gene Set Enrichment Analysis revealed a significant enrichment of the monocyte core gene signature within the expression profile of CD14+/lowSIRPα+CD103- cells, and the cDC2 signature was enriched in CD14-SIRPα+CD103+ cells, while all other comparisons did not reach statistical significance (not shown).
The calprotectin subunits S100A8 and S100A9 and Flt3 were contained in gene clusters 1 and cluster 2/3, respectively, and exhibited strongly dichotomous and reciprocal expression patterns; while CD14+ subsets including CD14+/low cells expressed high levels of S100A9 (Fig. 5d, upper panel) and S100A8 (Fig. 5d, middle panel) and low levels of FLT3, CD14- subsets showed high FLT3 expression and very low levels of both calprotectin subunits. In contrast, genes in clusters 2/3 such as CD5 correlated with Flt3 expression (Fig. 5d, lower panel).
Taken together, the results of the RNA-seq analysis strongly suggest that the Flt3-CD14+/low subsets are related to the monocyte lineage, while CD14- subsets are bona fide cDC2.
Antigen processing and migration capacity of CD14+/low SIRPα+ APCs resembles that of Mf
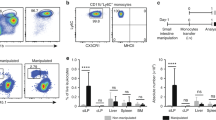
We next characterized the functional properties of the APC subsets in healthy, non-inflamed tissues, again using CD14-beads to separate monocyte-related cells and bona fide cDCs. A hallmark of cDCs is the capacity to capture antigens and migrate to local lymph nodes for T cell activation, whereas monocytes rapidly recruited to sites of infection are important for clearing pathogens and cellular debris.46
To assess the ability of the different APC subsets to process antigens we cultured cells with DQ-conjugated ovalbumin, DQ-BSA, or labeled particulate antigen pHrodo E.coli. The self-quenched DQ-conjugates are taken up by fluid phase endocytosis, emit pH-independent fluorescence in the green spectrum after proteolysis and red fluorescence when accumulating in lysosomes. The phagocytic and endocytic capacity of different hSI Mf subsets is higher than those of the overall DC population.31 We could define more precisely that intestinal CD14+CD11c+ Mf show significantly higher efficiency than CD14- DCs, but similar to CD14+/low SIRPα+ APCs. The latter subset exhibited a significantly higher processing capacity for either type of antigen compared to CD14- DCs (Fig. 6a). SIRPα- DCs showed very low processing capacity. Only 16.65 ± 8.55% (N = 3) of SIRPα- DCs had processed DQ-Ova even after prolonged incubation (data not shown). Although antigen uptake and processing capacity among blood DCs was generally lower than for mucosal DCs, the difference between cDC1 and cDC2 was also pronounced in blood (Supplementary Figure 7).
Antigen uptake and migration of CD14+/low subsets resembles those of Mf. a Flow cytometric quantification of processing of DQ-Ova (N = 6), DQ-BSA (N = 3), and particulate antigen pHrodo E.coli (N = 3) by Mf and DC subsets resulting from CD14-specific bead-based separation. Gates were set based on corresponding cells incubated with the substrates at 4 °C. a,b,c: data sets that do not share any indicated letters are significantly different from each other (RM-ANOVA). b Relative abundance of hSI DC subsets among live CD45+HLA-DRhiCD14-CD11c+ cells in fresh tissue (total), walk-out cells, and residual tissue cells (RM-ANOVA). c Abundance of the subsets among walk-out cells with/without CCL21 stimulation, normalized to abundance in fresh tissue. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001 (RM-ANOVA). d Gene expression data for CCR7 from RNA-seq data. * P < 0.05 (t-test, excluding SIRPα- and Mf). e Normalized CCR7 mRNA levels assessed by qPCR after 24 h culture of sorted tissue APC subsets with/without LPS stimulation. * P < 0.05 ** P < 0.01 (ANOVA and ANOVA with Tukey posthoc test for subset effects; multiple t-tests with Holm-Sidak correction for treatment effect within subset). For (d) and (e) tissue cells were sorted as described in Fig. 4c
To assess the migratory potential of the APC subsets we cultured biopsies of intestinal mucosa, and phenotyped leukocytes exiting the tissue. The SIRPα+CD103+ subset consistently showed the highest rates of spontaneous migration out of the intestinal tissue, while SIRPα+CD103- DCs appeared to be more stationary, and SIRPα-CD103+ cells showed an intermediate rate of spontaneous migration (Fig. 6b). Migration of DCs to draining lymph nodes is CCR7-dependent,2 and addition of the CCR7-ligand CCL21 increased the migratory capability in both CD103+ subsets, but not SIRPα+CD103- cells (Fig. 6c). Our data indicate that the latter subset consists to a large extent of calprotectin+CD14+/low monocyte-related cells. We therefore assessed CCR7 mRNA expression in the RNA-seq data of FACS-sorted subsets and CD14+CD11c+ Mf. Among the SIRPα+ subsets, CD14+/low exhibited significantly lower levels of CCR7 expression (Fig. 6d). In addition, we performed qPCR after culturing the FACS-sorted subsets with or without LPS for 24 h. Again, CD14- but not CD14+/low APCs showed higher expression levels of CCR7 compared to CD14+CD11c+ Mf irrespective of LPS stimulation (Fig. 6e). Addition of LPS significantly increased CCR7 expression in CD14-SIRPα+CD103+ DCs only (Fig. 6e). These findings strongly suggest that monocyte-related cells have a limited potential for CCR7-dependent migration.
Only CD14- Flt3+ SIRPα+ cDCs efficiently prime naïve CD4+ T cells
Although previously considered a unique ability of classical DCs, monocyte-derived cells have been shown to activate naïve T cells in mice.27 In order to assess this priming potential in steady-state hSI APCs, we again used CD14-beads to separate tissue monocyte-related cells and bona fide cDCs, and sorted the APC subsets as above for co-cultures with allogeneic naïve T cells from blood. CD14-SIRPα+CD103+ DCs and to a lesser extent CD14-SIRPα+CD103- induced proliferation of naïve CD4+ T cells and IL-2 secretion. SIRPα- DCs-stimulated CD4+ T cells showed some CD25 expression and IL-2 production, but very low proliferation levels (Fig. 7a). Of note, CD14+/lowCD103-SIRPα+ and CD14+/lowCD103+SIRPα+ cells and Mf were similarly poor inducers of CD4+ T cell proliferation (Fig. 7a). CD14-SIRPα+ DCs induced Th1 type T cells, expressing the transcription factor T-bet (Fig. 7b). Moreover, only co-culture supernatants of CD14-SIRPα+ DCs were enriched in the Th1 cytokines IFNγ and TNFα (Fig. 7c, d). IL-10 was only detected at low levels and IL-17 was undetectable in culture supernatants (data not shown). Intracellular staining after PMA/ionomycin stimulation revealed induction of IL-17A+ T cells exclusively after coculture with CD14-SIRPα+CD103+ DCs (Fig. 7e). Collectively, these data indicate that pre-cDC-derived hSI DCs, but not monocyte-related SIRPα+ cells efficiently drive proliferation and Th1/Th17 differentiation in naïve allogeneic CD4+ T cells.
Only Flt3+ DCs efficiently prime naïve T cells. a Flow cytometric quantification of proliferation (CFSE dilution) and CD25 expression in allogeneic CD4+ T cells after coculture with the different subsets of tissue DCs and CD14+CD11c+ Mf (N = 4). Tissue cells were sorted as described in Fig. 4c. IL-2 concentration in coculture supernatants from day 6 was analyzed by Bio-Plex Assay. (b) Abundance of T-bet+ and (e) IL17A+ CD4+ T cells after PMA/ionomycin stimulation. (c) IFNγ and (d) TNF concentrations in coculture supernatants were analyzed by Bio-Plex Assay (ANOVA)
Discussion
The origin and functional heterogeneity of human intestinal DC subsets is insufficiently understood. By assessing the expression of monocyte-specific markers and RNA-sequencing of phenotypically refined subsets we show here that monocyte-related cells contribute substantially to the cell compartment usually defined as DCs in steady-state hSI. These monocyte-related cells were both transcriptionally and functionally similar to hSI Mf.
Monocytes are highly versatile and can acquire diverse phenotypic and functional capabilities in response to environmental cues. The complexity of monocyte phenotypes during their differentiation in tissues previously prevented the implementation of an unambiguous nomenclature within the mononuclear phagocyte compartment. In fact, our data suggest an even closer phenotypic resemblance between blood monocytes, tissue monocyte-related cells, and bona fide cDC2 than previously appreciated, as all subsets express SIRPα, and are able to express CD103, CD1c,31,47 and other markers previously used to define cDC2.
Lineage-specifying transcription factors are obvious identifiers of the respective populations, but the newly described markers for monocyte-related cells such as S100A-proteins or TREM1 might prove applicable in the future due to their higher protein expression levels. Interestingly, the set of genes we found to characterize monocyte-related cells mimicking DC phenotype in hSI are somewhat overlapping (calprotectin, CD14, TREM) with genes defining a recently described subset of blood cDC2.48 However, this studied RNAseq dataset stems from one donor only, and cDC2 heterogeneity is likely highly variable across individuals,49 precluding a direct comparison with our datasets.
Based on expression of the marker calprotectin we estimate that about half of all SIRPα+ APCs are Flt3- and show a monocyte-related transcriptional signature. However, as this study and previously published data demonstrate that the recruited blood monocytes down-regulate intracellular calprotectin in the tissue,31,39,43 our data might underestimate the quantitative contribution of monocyte-related cells to the compartment of SIRPα+ DCs.
According to our data, monocyte-related cells and bona fide DCs exhibit distinct functional specialization despite their phenotypic similarities. In vivo migration to MLNs is challenging to assess in humans, in particular since activation, differentiation and migration affect the expression of cell surface markers, such as chemokine receptors, for the distinction of cell subsets. Our transcriptional profiling and functional studies suggest that monocyte-related cells have limited migratory potential, concordant with mouse data, showing that primarily pre-cDC-derived cells efficiently migrate from peripheral tissues during steady-state and following antigen exposure.24,35 The monocyte-related CD14+/lowSIRPα+ subset was very efficient in antigen uptake but failed to efficiently prime naïve T cells. This is in line with reports that monocytes and monocyte-related cells exhibit low capacity to prime naïve CD4+ T cells in both humans and mice.16,34,50 Our data confirm earlier descriptions of the migratory potential of hSI DC subsets,16,51 and reveal characteristics of the described populations in the steady-state, which might be altered in an inflammatory environment.
To our knowledge, we here provide the first assessment of the in vivo turnover of hSI cDCs. Although we found a substantial number of cells potentially proliferating in situ, in particular among calprotectin- cDCs, our data demonstrate a virtually complete replacement of the DC population within 6 weeks. This rapid turnover of human DC contrasts the longevity of other human tissue APCs such as Langerhans cells,52,53 and alveolar Mf,43 but is in concordance with mouse studies describing a maintenance of the cDC population in lymphoid and peripheral tissues by both in situ proliferation and constant replenishment from blood-borne precursors, with complete replacement of the DC compartment within 14 days.15,54,55,56 While only transplanted patients without signs of rejection episodes were considered, we cannot exclude that the replacement rate was affected by a low-grade allograft inflammatory response. However, CD11c- Mfs residing in the same tissue specimen survived for > 6 weeks,31 while memory B cells and antibody-secreting plasma cells persisted for > 1 year.57
Collectively, our findings provide evidence for a significant contribution of monocyte-related cells to the compartment of SIRPα+ APCs usually gated as DCs in the hSI, exerting distinct functions to sustain intestinal homeostasis. In a broader context, our data highlight the diverseness of tissue monocyte-derived cells, as well as the ontogenetic and functional heterogeneity of the compartment that is typically defined as DCs based on phenotype. This contribution to the understanding of the ontogeny of mononuclear phagocyte subsets in human tissues will further add to the ongoing debate about terminology, and underscores that a logical nomenclature within this compartment should be based on ontogeny rather than mere phenotypic characterization. The study also provides a framework for further investigation of which (micro-)environmental cues determine whether monocytes entering the intestinal mucosa differentiate into classical Mf or cells mimicking a DC phenotype.
Methods
Samples and cell isolation
DCs were isolated from the proximal jejunum resected during Whipple procedure on pancreatic cancer patients at Oslo University Hospital, Rikshospitalet. In total, 105 tissues from 52 males (mean age 68, range 52–82), 48 females (mean age 72, range 53–89) and 5 donors with unknown gender were analyzed. The resected intestine (5–10 cm) was opened longitudinally and washed in PBS. Mucosal folds were excised and epithelial cells were removed by three washing steps with 2 mM EDTA in PBS for 20 min at 37 °C. The remaining LP was digested in RPMI containing 0.25 mg/ml liberase (Roche) and 20 U/ml DNase I (Roche) at 37 °C. Mononuclear cells were enriched by lymphoprep™ (Stemcell technologies) density gradient centrifugation. Histological evaluation was performed blindly by an experienced pathologist based on routinely H&E stained formalin-fixed tissue specimen. Most samples showed normal architecture with no signs of inflammation, but in some cases low-grade inflammation with the presence of neutrophils and an increased density of leukocytes were found without any known cause.
Blood samples were obtained from healthy adult blood donors at Ullevål University Hospital or Rikshospitalet. Peripheral blood mononuclear cells were isolated by lymphoprep™ density gradient centrifugation.
Replacement of donor monocytes/DCs by recipient cells was assessed in transplanted duodenum in type I diabetes patients that had received pancreas-duodenum transplantation as described in detail previously.40 Biopsies of the grafted duodenum were obtained 3 and 6 weeks after transplantation. Histological evaluation was performed blindly by experienced pathologists and only biopsies from patients without signs of acute rejection58 and HLA class I-mismatch were included. HLA-mismatch enabled us to distinguish donor and recipient cells by flow cytometry of single cell suspension of LP cells. Stromal cells were analyzed as intrinsic control for the precision of the sampling.
All participants provided written consent, and the study was approved by the Norwegian Regional Committee for Medical Research Ethics (2012/2278/REK sørøst and 2010/2720/REK sørøst; Biobank: “Tarmsykdommer” REK 2012/341).
See Supplementary Methods for details on flow cytometry procedures, antibodies, RNA sequencing and analysis of RNA-seq data, functional studies and statistics. Annotated representative flow cytometry data are publicly available through FlowRepository.org. Data were acquired on a BD LSR Fortessa or BD FACS ARIA II and analyzed using FlowJo software (FlowJo, LLC; now BD). Shaded areas represent FMO controls (surface antigens) or matched isotype-control antibody staining (intracellular antigens).
References
Banchereau, J. & Steinman, R. M. Dendritic cells and the control of immunity. Nature 392, 245–252 (1998).
Lanzavecchia, A. & Sallusto, F. Regulation of T cell immunity by dendritic cells. Cell 106, 263–266 (2001).
Fogg, D. K. et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 311, 83–87 (2006).
Lee, J. et al. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J. Exp. Med. 212, 385–399 (2015).
Naik, S. H. et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8, 1217–1226 (2007).
McKenna, H. J. et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood 95, 3489–3497 (2000).
See, P., et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science 356, 6342, eaag3009 (2017).
Breton, G. et al. Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs. J. Exp. Med. 213, 2861–2870 (2016).
Schlitzer, A. et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 16, 718–728 (2015).
Guilliams, M. et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol. 14, 571–578 (2014).
Schiavoni, G. et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J. Exp. Med. 196, 1415–1425 (2002).
Hildner, K. et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008).
Schlitzer, A. et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity 38, 970–983 (2013).
Suzuki, S. et al. Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha- dendritic cell development. Proc. Natl. Acad. Sci. USA 101, 8981–8986 (2004).
Liu, K. et al. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat. Immunol. 8, 578–583 (2007).
Watchmaker, P. B. et al. Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat. Immunol. 15, 98–108 (2014).
Persson, E. K. et al. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 38, 958–969 (2013).
Edelson, B. T. et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med. 207, 823–836 (2010).
Haniffa, M. et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity 37, 60–73 (2012).
Crozat, K. et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J. Exp. Med. 207, 1283–1292 (2010).
Poulin, L. F. et al. DNGR-1 is a specific and universal marker of mouse and human Batf3-dependent dendritic cells in lymphoid and nonlymphoid tissues. Blood 119, 6052–6062 (2012).
Ginhoux, F. et al. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206, 3115–3130 (2009).
Schulz, O. et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 206, 3101–3114 (2009).
Schreiber, H. A. et al. Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. J. Exp. Med. 210, 2025–2039 (2013).
Sheng, J. et al. A discrete subset of monocyte-derived cells among typical conventional type 2 dendritic cells can efficiently cross-present. Cell Rep. 21, 1203–1214 (2017).
Scott, C. L. et al. CCR2(+)CD103(-) intestinal dendritic cells develop from DC-committed precursors and induce interleukin-17 production by T cells. Mucosal Immunol. 8, 327–339 (2015).
Zigmond, E. et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 37, 1076–1090 (2012).
Tamoutounour, S. et al. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur. J. Immunol. 42, 3150–3166 (2012).
Melum, G. R. et al. A thymic stromal lymphopoietin-responsive dendritic cell subset mediates allergic responses in the upper airway mucosa. J. Allergy Clin. Immunol. 134, 613–621 e617 (2014).
Mair, F. & Prlic, M. OMIP-044: 28-color immunophenotyping of the human dendritic cell compartment. Cytom. A 93, 402–405 (2018).
Bujko, A. et al. Transcriptional and functional profiling defines human small intestinal macrophage subsets. J. Exp. Med. 215, 441–458 (2017).
Dai, X. M. et al. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99, 111–120 (2002).
Cecchini, M. G. et al. Role of colony stimulating factor-1 in the establishment and regulation of tissue macrophages during postnatal development of the mouse. Development 120, 1357–1372 (1994).
Bain, C. C. et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 6, 498–510 (2013).
Plantinga, M. et al. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity 38, 322–335 (2013).
Seo, S. U. et al. Intestinal macrophages arising from CCR2(+) monocytes control pathogen infection by activating innate lymphoid cells. Nat. Commun. 6, 8010 (2015).
Odink, K. et al. Two calcium-binding proteins in infiltrate macrophages of rheumatoid arthritis. Nature 330, 80–82 (1987).
Rugtveit, J., Brandtzaeg, P., Halstensen, T. S., Fausa, O. & Scott, H. Increased macrophage subset in inflammatory bowel disease: apparent recruitment from peripheral blood monocytes. Gut 35, 669–674 (1994).
Rugtveit, J., Scott, H., Halstensen, T. S., Norstein, J. & Brandtzaeg, P. Expression of the L1 antigen (calprotectin) by tissue macrophages reflects recent recruitment from peripheral blood rather than upregulation of local synthesis: implications for rejection diagnosis in formalin-fixed kidney specimens. J. Pathol. 180, 194–199 (1996).
Horneland, R. et al. Pancreas transplantation with enteroanastomosis to native duodenum poses technical challenges--but offers improved endoscopic access for scheduled biopsies and therapeutic interventions. Am. J. Transplant. 15, 242–250 (2015).
McGovern, N. et al. Human dermal CD14(+) cells are a transient population of monocyte-derived macrophages. Immunity 41, 465–477 (2014).
Haniffa, M. et al. Differential rates of replacement of human dermal dendritic cells and macrophages during hematopoietic stem cell transplantation. J. Exp. Med. 206, 371–385 (2009).
Eguiluz-Gracia, I. et al. Long-term persistence of human donor alveolar macrophages in lung transplant recipients. Thorax 71, 1006–1011 (2016).
Segura, E., Durand, M. & Amigorena, S. Similar antigen cross-presentation capacity and phagocytic functions in all freshly isolated human lymphoid organ-resident dendritic cells. J. Exp. Med. 210, 1035–1047 (2013).
Frankenberger, M. et al. Transcript profiling of CD16-positive monocytes reveals a unique molecular fingerprint. Eur. J. Immunol. 42, 957–974 (2012).
Shi, C. & Pamer, E. G. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 11, 762–774 (2011).
Schroder, M. et al. CD1c-Expression by monocytes—implications for the use of commercial CD1c+ dendritic cell isolation kits. PLoS ONE 11, e0157387 (2016).
Villani, A. C., et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, 6335, eaah4573 (2017).
Alcantara-Hernandez, M. et al. High-dimensional phenotypic mapping of human dendritic cells reveals interindividual variation and tissue specialization. Immunity 47, 1037–1050 e1036 (2017).
Rivollier, A., He, J., Kole, A., Valatas, V., & Kelsall, B. L. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J. Exp. Med. 209, 139–155 (2012).
Al-Hassi, H. O. et al. A mechanistic role for leptin in human dendritic cell migration: differences between ileum and colon in health and Crohn’s disease. Mucosal Immunol. 6, 751–761 (2013).
Kanitakis, J., Petruzzo, P., & Dubernard, J. M. Turnover of epidermal Langerhans’ cells. N. Engl. J. Med. 351, 2661–2662 (2004).
Merad, M. et al. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat. Immunol. 3, 1135–1141 (2002).
Kabashima, K. et al. Intrinsic lymphotoxin-beta receptor requirement for homeostasis of lymphoid tissue dendritic cells. Immunity 22, 439–450 (2005).
Diao, J. et al. In situ replication of immediate dendritic cell (DC) precursors contributes to conventional DC homeostasis in lymphoid tissue. J. Immunol. 176, 7196–7206 (2006).
Ruedl, C., Koebel, P., Bachmann, M., Hess, M. & Karjalainen, K. Anatomical origin of dendritic cells determines their life span in peripheral lymph nodes. J. Immunol. 165, 4910–4916 (2000).
Landsverk, O. J. et al. Antibody secreting plasma cells persist for decades in human intestine. J. Exp. Med. 214, 309–317 (2017).
Ruiz, P. et al. International grading scheme for acute cellular rejection in small-bowel transplantation: single-center experience. Transplant. Proc. 42, 47–53 (2010).
Acknowledgements
The authors thank Kathrine Hagelsteen, Kjersti Thorvaldsen Hagen, and Aaste Aursjø for technical assistance; the staff at the Endoscopy Unit, the surgery theatre, and Raquel Bartolomé for help with sample preparation; the Flowcytometry Core Facility at Oslo University Hospital Rikshospitalet, the Department for Microbiology, Oslo University Hospital Rikshospitalet, and the Core Facility Flow Cytometry at Biomedical Center, Ludwig-Maximilians-University Munich, for sharing equipment. This work was partly supported by the Research Council of Norway through its Centres of Excellence funding scheme, project number 179573/V40; and by grant from the South Eastern Norway Regional Health Authority, project number 1012105.
Author information
Authors and Affiliations
Contributions
L.R., E.S.B. and F.L.J. conceived the study. L.R. performed experiments and wrote the manuscript. O.J.B.L. performed experiments and edited the manuscript. A.B. and N.A. performed experiments and established methodology. S.Y. coordinated recruitment and collection of clinical tissue samples. R.H., O.Ø., and E.M.Aa. were responsible for recruitment and clinical follow-up of the transplanted patients. K.E.A.L was responsible for endoscopic examination, biopsy procedures, and the biobank. H.G.S., E.S.B., and F.L.J. supervised the study and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Richter, L., Landsverk, O.J.B., Atlasy, N. et al. Transcriptional profiling reveals monocyte-related macrophages phenotypically resembling DC in human intestine. Mucosal Immunol 11, 1512–1523 (2018). https://doi.org/10.1038/s41385-018-0060-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-018-0060-1
This article is cited by
-
Genetic models of human and mouse dendritic cell development and function
Nature Reviews Immunology (2021)
-
Response to Lauro and Zorzetti
Mucosal Immunology (2021)