Abstract
Blood vessel epicardial substance (BVES), or POPDC1, is a tight junction-associated transmembrane protein that modulates epithelial-to-mesenchymal transition (EMT) via junctional signaling pathways. There have been no in vivo studies investigating the role of BVES in colitis. We hypothesized that BVES is critical for maintaining colonic epithelial integrity. At baseline, Bves−/− mouse colons demonstrate increased crypt height, elevated proliferation, decreased apoptosis, altered intestinal lineage allocation, and dysregulation of tight junctions with functional deficits in permeability and altered intestinal immunity. Bves−/− mice inoculated with Citrobacter rodentium had greater colonic injury, increased colonic and mesenteric lymph node bacterial colonization, and altered immune responses after infection. We propose that increased bacterial colonization and translocation result in amplified immune responses and worsened injury. Similarly, dextran sodium sulfate (DSS) treatment resulted in greater histologic injury in Bves−/− mice. Two different human cell lines (Caco2 and HEK293Ts) co-cultured with enteropathogenic E. coli showed increased attaching/effacing lesions in the absence of BVES. Finally, BVES mRNA levels were reduced in human ulcerative colitis (UC) biopsy specimens. Collectively, these studies suggest that BVES plays a protective role both in ulcerative and infectious colitis and identify BVES as a critical protector of colonic mucosal integrity.
Similar content being viewed by others
Introduction
Inflammatory bowel disease (IBD) affects over 1.6 million people in the United States, causing numerous and debilitating complications such as malnutrition, infection, pain, and an elevated risk of colorectal cancer.1,2 The fundamental pathophysiology of IBD is not well understood. Intestinal mucosal homeostasis relies on continual proliferation, differentiation, and repair of the epithelium, and these processes are challenged in response to injury.3,4,5 Maintenance of intestinal integrity requires competent epithelial barrier function as well as intact repair programs to respond to intestinal challenges.3,4,5 The epithelial barrier is “leaky” in patients with IBD; multiple studies demonstrate lower epithelial resistance and increased permeability of both inflamed and non-inflamed mucosa in Crohn’s disease (CD) and ulcerative colitis (UC).6,7 Tight junctions are apical complexes critical to the maintenance of epithelial barriers.8,9 Disruption of tight junctional complexes is a key contributing factor to epithelial barrier dysfunction.6,7,10 For example, tight junction-associated proteins, such as F11r, which we will refer to as junctional adhesion molecule-A (JAM-A), and MYH2, which we will refer to as non-muscle myosin II (NMII), are protective in the dextran sodium sulfate (DSS) model of experimental colitis.11,12 Loss of either JAM-A11 or NMII12 resulted in low-grade inflammation and increased intestinal permeability, but interestingly, the mice were able to maintain intestinal homeostasis and were otherwise normal at baseline. However, upon colitogenic challenge, these mice were unable to compensate, resulting in markedly more severe injury.
Blood vessel epicardial substance (BVES) is a tight junction-associated, three-pass transmembrane protein originally discovered from a cDNA screen of the developing heart.13,14 BVES is expressed in epithelial tissues, including the intestinal epithelium, and regulates epithelial-to-mesenchymal transition (EMT).13,15,16,17,18,19,20 We have previously demonstrated that BVES regulates stem cell programs in the small intestine,20 colonic epithelial phenotypes in vitro,15 and can promote inflammatory tumorigenesis in vivo,21 but its role in colitis remains unexplored. We hypothesized that BVES is critical for maintaining epithelial integrity after colonic injury.
At baseline, Bves−/− mice exhibited higher colonocyte proliferation, decreased apoptosis, defects in intestinal secretory lineage allocation, and dysregulation of tight junction proteins. Ex vivo studies, electron microscopy (EM), and 2D colonoid cultures demonstrated increased permeability in Bves−/− colons. Moreover, Bves−/− mice displayed dysregulated mucosal chemokine and cytokine production. After inoculation with Citrobacter rodentium, Bves−/− mice had exacerbated colitis along with increased colonic and mesenteric lymph node bacterial colonization, decreased proliferation and stem cell marker expression, and skewing of mucosal chemokines and cytokines. In confirmatory studies, Bves−/− mice treated with DSS also had increased histologic injury in both acute and injury-repair models. BVES silencing in HEK293T and Caco2 cells resulted in increased attaching/effacing lesions and CXCL8 mRNA expression after EPEC infection, while HEK293T cells overexpressing BVES had fewer attaching/effacing lesions and decreased mRNA expression of CXCL8. Lastly, analysis of human UC mucosal biopsies demonstrated decreased BVES expression, suggesting that its downregulation is relevant to intestinal injury processes. Collectively, these findings indicate that BVES regulates the mouse colonic microenvironment, protecting against intestinal epithelial injury and inflammation, and may play a protective role in IBD.
Materials And Methods
Mouse models
Wild-type mice (WT; C57BL/6 background) were obtained from Jackson Laboratories. Bves−/− mice have previously been described in detail.15 All experiments were performed with 8–12-week-old male and female mice on C57BL/6 background under guidelines approved by the Vanderbilt University Institutional Animal Care and Use Committee.
Functional permeability analysis
These experiments were conducted as previously described.22 Briefly, sections of distal colon from 8–12-week-old male mice were mounted between the two halves of the Ussing Chamber (1.12 cm2 opening). The mucosal and serosal sides of chambers were connected to circulating reservoirs that contained Ringer’s solution with 5 mM mannitol. A gas lift column of 95% O2 maintained oxygenation and pH 7.4 of reservoirs. Isc was continuously monitored on a strip chart recorder, and electrical resistance was calculated using Ohm’s law. After baseline potential difference and resistance was measured, a fluorescent tracer (FITC–dextran, 4 kDA, #46944, Sigma) at a concentration of 80 mg/ml was placed on one side (luminal bath) and concentration was measured 3 h after beginning experiment on the other side (serosal bath) via spectrophotofluorometry, with an excitation wavelength of 492 nm and emission wavelength of 518 nm.
Electron microscopy
Specimens were processed for transmission electron microscopy (TEM) and imaged in the Vanderbilt Cell Imaging Shared Resource-Research Electron Microscopy facility.
Embedding
Samples were fixed in 2.5% gluteraldehyde in 0.1 M cacodylate buffer, pH 7.4 at room temperature (RT) for 1 h and then transferred to 4 °C, overnight. The samples were washed in 0.1 M cacodylate buffer, then incubated 1 h in 1% osmium tetraoxide at RT, and then washed with 0.1 M cacodylate buffer. Subsequently, the samples were dehydrated through a graded ethanol series and then three exchanges of 100% ethanol. Next, the samples were incubated for 5 min in 100% ethanol and propylene oxide (PO), followed by two exchanges of pure PO. Samples were then infiltrated with 25% Epon 812 resin and 75% PO for 30 min at RT. Next, they were infiltrated with Epon 812 resin and PO [1:1] for 1 hour at RT then stored overnight at RT. The next day the samples went through a [3:1] (resin:PO) exchange for 3–4 h and were incubated with pure epoxy resin overnight. Samples were then incubated in two more changes of pure epoxy resin and allowed to polymerize at 60 °C for 48 h.
Sectioning and imaging
Sections of 500–1000 nm thickness were cut for ultrastructure identification. Then, 70–80 nm ultrathin sections were cut from the region of interest and collected on 300-mesh copper grids and post-stained with 2% uranyl acetate and then with Reynold’s lead citrate. Samples were subsequently imaged on the Philips/FEI Tecnai T12 electron microscope at various magnifications.
Colonoid transepithelial resistance measurements
Colonoid cultures were derived according to previously published protocols.23 Briefly, the colon was dissected, flushed with ice-cold phosphate buffered saline (PBS), and opened lengthwise. Colons were cut into 5 mm pieces before being transferred to chelation buffer (2 mM EDTA in Dulbecco’s PBS) and rocked for 90 min at 4 °C. After removing the chelation buffer, cold shaking buffer (PBS with 43.3 mM sucrose and 54.9 mM sorbitol) was added and tissues were gently shaken for 4 min to free colonic crypts. Colonic crypts were plated in growth factor-reduced Matrigel (Corning, 356231) at a concentration of 25 crypts per 5-μL plug. After polymerization, Matrigel was overlaid with 500 μL mini-gut media (Advanced DMEM/F12, B27, N2, Pen-Strep, L-glutamine, HEPES) and supplemented with EGF (R&D systems, 2028-EG-200), Noggin (R&D Systems, 1967-NG-025/CF), and R-spondin (Vanderbilt Antibody and Protein Resource). For transwell plating, colonoids were collected and dissociated using TrypLE Express (Gibco, Life Technologies, 12604-013). Following dissociation, cells were plated on 0.4 µm transwell filters (Corning, 3413) coated with Collagen 1 (Gibco, A10483-01) and cultured in mini-gut media supplemented with 10 µM Y27632 (R&D Systems, 1254/10). After 1 day, mini-gut media was replaced by Advanced DMEM/F12 containing 1% penicillin/streptomycin, 1 × Glutamax (Gibco, Life Technologies, 35050-061), and 10% FBS. Transepithelial resistance (TER) was measured by Ohm meter daily for 5 days.
Western blot analysis
For western blot analysis, crypts were isolated by the same method for colonoid culture described above. Crypts were pelleted by centrifugation and resuspended in RIPA buffer, phosphatase inhibitor cocktail 2 (Sigma, P5726), phosphatase inhibitor cocktail 3 (Sigma, P0044), protease inhibitor (Sigma, P8340), and 2 × Laemmli buffer and then boiled for 5 min before analysis by SDS-PAGE electrophoresis. Membranes were blocked using Odyssey blocking buffer for 30 min and then blotted with anti-E-cadherin (BD Transductions, #610182), anti-ZO-1 (Life Technologies, #617300), anti-Claudin 7 (Invitrogen #QL221418), anti-GAPDH (Cell Signaling, #5174S), anti-β-actin (Sigma, #116M4801V), anti-myosin light chain 2 (Cell Signaling, #8505S), anti-phospho-myosin light chain 2(Cell Signaling, #3674S), or Myosin 2 C (Cell Signaling, #8189S). All antibodies were used at 1:1000 concentration (except anti-β-actin, 1:2000) in Odyssey blocking buffer with 0.1% Tween-20 and incubated overnight at 4 °C. LiCor secondary antibodies were used at 1:10,000 dilution and incubated for 2 h at RT. Quantification of western blot band intensity was conducted using LiCor Image Studio.
Induction of Citrobacter rodentium colitis
Mice were orally inoculated with C. rodentium as done previously.24,25,26,27 Bacteria were grown overnight in Luria broth (LB), and subculture was performed the following day to ensure live bacteria were in exponential phase. Mice were infected by oral gavage with 0.1 mL of broth containing 5 × 108 CFU of C. rodentium. Subculture was performed the following day to ensure that live bacteria were in exponential phase. Control mice received sterile broth. At 14 days post inoculation, the animals were killed and the colons were removed, cleaned, and Swiss-rolled for histology. For colonization studies, distal colon pieces or mesenteric lymph nodes were homogenized, serially diluted in LB, and plated on MacConkey agar plates. For colonic tissues, colonies were counted after 24 h and CFU/g was calculated. For mesenteric lymph nodes, colonies were counted after 48 h and CFU/g was calculated. The C. rodentium histology slides were reviewed and scored in a blinded manner by a gastrointestinal pathologist (M.B.P.). Both acute (neutrophilic) and chronic (lymphocytic) inflammation, extent of inflammation, and epithelial damage were each scored on 0–3 scale.24,27 The aggregate histologic injury score was the sum of acute and chronic inflammation multiplied by the extent of inflammation, plus the epithelial injury (0–21).27
Immunohistochemistry
For histological assessment, colons were removed, Swiss-rolled, and fixed in 10% formalin overnight prior to submission to the Vanderbilt University Translational Pathology Shared Resource (TPSR) core for processing and paraffin-embedding. For immunohistochemistry (IHC), 5 µm sections were cut, dewaxed, hydrated, and endogenous peroxidase activity quenched with 0.03% hydrogen peroxide in MeOH.28,29,30 Antigen Unmasking Reagent (Vector Laboratories, H-3300) was used for antigen retrieval. The Vectastain ABC Elite System (Vector Laboratories, AK-5002) was then used according to the manufacturer’s instructions. Primary antibodies were added overnight at 4 °C with isotype-matched antibodies (negative controls) on serial sections. The following antibodies were used: anti-phospho-Histone H3 (pH3) Ser10 antibody (1:150, #2465253, Millipore), anti-CgA (1:1000, #280170, ImmunoStar Inc.), an anti-CD3 antibody (1:200, MCA500GA/1110, Bio-Rad). Periodic Acid Schiff (PAS) staining for goblet cells and hematoxylin and eosin (H&E) staining was performed by the TPSR core. Apoptotic cells were identified using the ApopTag Plus Peroxidase In Situ Apoptosis Kit (Millipore, S7101) according to the manufacturer’s protocol. Quantification was generated by counting cells in 100 sequential, well-aligned crypts from the distal colon and presented as the mean number of positive cells per crypt ± SE.
For fluorescent IHC on paraffin-embedded sections, antigen retrieval consisted of 10 min boiling in 10 mM sodium citrate (pH 6). After blocking, samples were incubated with the following primary antibodies overnight at 4 °C: anti-Ly6B.2 antibody (1:100, Bio-Rad, MCA771G), anti-B220 antibody (1:500, BD Pharmingen, 550286), anti-E-cadherin (1:500, #610182, BD Transduction Laboratories), or anti-ZO-1 (1:500, Life Technologies, #617300). Secondary antibodies were conjugated to either 488 or 594 Alexafluor dyes (1:1000, Invitrogen) and incubated for 2 h at RT. Nuclear staining was done with ProLong Gold antifade reagent and DAPI (Invitrogen, 1846939).
For fluorescent IHC on frozen sections, colons were removed, Swiss-rolled, and immediately frozen in optimal cutting temperature (OCT) compound (Tissue-Tek, #4583). Five μm sections of OCT-embedded tissue were cut using a cryostat. Sections were fixed with 1% paraformaldehyde, permeabilized with Triton X-100, and blocked/quenched with 10% normal goat serum (50% mM HN4Cl-filtered). Samples were then incubated with the following primary antibodies overnight at 4 °C: anti-phospho-myosin light chain 2 (1:200, Cell Signaling, 3674S) and Myosin IIc (1:200, Cell Signaling, 8189S), Myosin IIb (1:200, Cell Signaling, 8824 T), and Phalloidin (1:500, Biotium, 00043 T). Secondary antibodies were conjugated to either 488 or 594 Alexafluor dyes (1:1000, Invitrogen) and incubated for 2 h at RT. Nuclear staining was done with ProLong Gold antifade reagent and DAPI (Invitrogen, 1846939).
qRT-PCR analysis
RNA from Bves−/− or WT colons was isolated using the Direct-zol RNA MiniPrep Kit (Genesee Scientific, #11-311) or RNeasy Mini Kit (Qiagen, #74106), and cDNA was synthesized using the qScript cDNA synthesis kit (Quantabio, #95047-500) or iScript cDNA synthesis kit (Bio-rad, #1708891) from 1 µg of total RNA. cDNA (1 µL) was used as a template in each subsequent PCR reaction. Taqman probes were purchased from Applied Biosystems, and CXCL8/IL-8 (Hs00174103) and BVES (Hs00362584) were analyzed using the delta–delta Ct method and normalized to GAPDH (Hs02758991).
Chemokine and Cytokine profiling
Fresh excised colon was rinsed and homogenized in CelLytic MT Mammalian Tissue Lysis/Extraction buffer (Sigma, St Louis, MO). Colonic tissue lysates were analyzed using the 25-analyte MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel (EMD Millipore) on a FLEXMAP 3D instrument. Protein concentration was measured using the Bio-Rad DC protein assay kit (Bio-Rad Laboratories). Concentration of each chemokine or cytokine was presented as pg/mg colon tissue protein.
DSS treatment
Acute DSS injury
Eight- to twelve-week-old WT and Bves−/− C57BL/6 mice were treated with 4% DSS (Affymetrix, #14489) in the drinking water for 5 days and then killed. Colonoscopy (Karl Storz, Veterinary Endoscopy) was performed before being killed and videos were scored using the murine endoscopic index of colitis severity (MEICS),31 which is a 15-point score based on thickening of the colon (0–3), changes of the vascular pattern (0–3), fibrin visibility (0–3), granularity of the mucosal surface (0–3), and stool consistency (0–3). After being killed, colons were Swiss-rolled and analyzed for histologic injury in a blinded manner by a gastrointestinal pathologist (M.K.W.) via calculation of an ulceration score, as previously described.32 Briefly, the scoring system utilized a score of 0–4 to grade the extent of destruction of the crypt architecture. A score of 0 indicated normal crypt architecture, 1 indicated effacement of the bottom third of the crypt, 2 indicated effacement of the bottom 2/3 of the crypt, 3 indicated complete effacement of the crypt, and 4 indicated complete crypt architectural effacement along with an absence of the normal epithelial surface.
DSS injury-repair model
Eight- to twelve-week-old WT and Bves−/− C57BL/6 mice were treated with 4% DSS for 5 days, allowed to recover for 5 days, and then killed. As in the acute DSS model, colons were Swiss-rolled and histologic injury calculated based on ulceration score.
Chronic DSS model
Eight- to twelve-week-old WT and Bves−/− C57BL/6 mice were treated with 4% DSS for 5 days and then allowed to recover for 10 days. This was repeated two additional times. Again, colons were Swiss-rolled and histologic injury calculated as described above.
Cell culture
HEK293T and Caco2 cells were grown in DMEM containing 10% FBS, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Polyethylenimine (PEI) at a concentration of 1 μg/mL was used for all transfection experiments. An empty vector was used to ensure equal quantities of cDNA were transfected. BVES was knocked down using siRNA (Santa Cruz, #60295). siRNA was transfected using Lipofectamine RNAiMAX (Invitrogen, #13778-150). Scrambled siRNA (Santa Cruz, #37007) was used as control.
Plasmids
BVES expression plasmids were generated using Gateway cloning (Invitrogen). pENTR vectors containing BVES (HsCD00368575) were shuttled into pcDNA 3.1V5 using lR clonase (Invitrogen).
EPEC infection/FAS test
EPEC strain E2348/69 was maintained on Luria-Bertani agar plates. The bacterial concentration was estimated to be 1 × 108 bacteria/ml per OD unit at 600 nm, as calculated by plating. HEK293T cells were plated on coverslips and infected with EPEC at a multiplicity of infection (MOI) of 10 for 3 h. Cells were washed with PBS, fixed with 3% paraformaldehyde, permeabilized in 0.1% Triton X, and stained with an actin probe (ActinGreen 488, Molecular Probes, #1608515) for 30 min. Coverslips were mounted on glass slides with ProLong Gold antifade reagent with DAPI (Invitrogen, 1846939) and visualized with a Zeiss LSM880 confocal microscope. Fifteen to twenty photos of high-powered fields (×63 or ×80) were taken and the number of attaching/effacing lesions were quantified by a blinded observer.
RNA Scope
High-resolution RNA in situ hybridization was performed using the RNAscope® 2.5 HD Detection Kit – BROWN according to the manufacturer’s protocol (ACDBio, document #322452 and #322310). Antigen retrieval was performed under standard pretreatment conditions as specified by the manufacturer. Probes were directed against BVES (catalog #434599), MsPPIB (positive control, catalog #313911), and DapB (negative control, catalog #310043).
Human subjects
Tissues were obtained during colonoscopies at the Vanderbilt University Medical Center, following a protocol approved by the Vanderbilt Institutional Review Board.28
Statistical Methods
Analyses comparing two groups were analyzed using the Man–Whitney non-parametric test. Kruskall–Wallis was used to compare multiple groups. Survival curve analysis was done using the Log-rank (Mantel–Cox) test. Data are presented as the mean ± SEM in bar graphs and a line identifying the mean is shown when all data points are plotted. All of these analyses were performed using GraphPad Prism®7.0c (San Diego, CA, USA). P < 0.05 was considered statistically significant.
Results
BVES regulates colonic crypt homeostasis
While it is known that loss of BVES increases susceptibility to colitis-associated carcinoma21 and regulates EMT,15 a process critical in injury-repair programs of the intestine,33,34 the impact of its deletion on colonic homeostasis in vivo has not previously been examined. Bves messenger RNA is detectable in the colonic epithelium and is distributed uniformly from the base to luminal surface of the crypt in WT mice (Supplementary Figure 1). To determine whether BVES deletion alters colonic phenotypes, we histologically characterized colons obtained from Bves−/− mice. Morphometric analysis demonstrated that Bves−/− mice had significantly greater crypt height (Fig. 1a). In determining the basis for this phenotype, we noted via TUNEL staining that crypt apoptosis was decreased (Fig. 1b) and phospho-histone H3 IHC revealed that proliferation was augmented in Bves−/− mice (Fig. 1c), suggesting an imbalance in crypt growth/clearance kinetics, which may contribute to the thickened mucosa observed in these mice. Abnormalities were not limited to growth parameters, as lineage allocation analysis revealed that the numbers of PAS-labeled goblet cells (Fig. 1d) and Cga-labeled enteroendocrine cells (Fig. 1e) were increased as compared to WT mice. Thus, BVES regulates colonic proliferation, apoptosis, lineage allocation, and crypt morphology, indicating a previously unrecognized role for BVES in influencing intestinal homeostasis.
BVES regulates colonic proliferation, lineage allocation, and crypt morphology. Colons were isolated and Swiss-rolled. a Representative H&E staining of sections of WT and Bves−/− colons. Images (left) and quantification (right) of WT and Bves−/− crypt height (201.7 μm vs. 251.2 μm, ***P < 0.001, n = 10). b Images (left) and quantification (right) of apoptotic cells per crypt unit (0.3 vs. 0.2 TUNEL+ cells/crypt, *P = 0.05, n = 8). c Images (left) and quantification (right) of crypt proliferation (0.6 vs. 1.5 phospho-Histone H3+ cells/crypt, **P < 0.01, n = 8). d Images (left) and quantification (right) of goblet cells/crypt (10.1 vs. 17.7 PAS+ cells/crypt, **P < 0.01, n = 6). e Images (left) and quantification (right) of enteroendocrine cells/crypt-villus unit (0.5 vs. 0.8 CgA+ cells/crypt, **P < 0.01, n = 5). All images were captured at ×20 magnification. Black arrows indicate positively stained cells in (b, e). Scale bars are in the bottom right corner of each image and are 100 μM in length
BVES contributes to epithelial barrier function
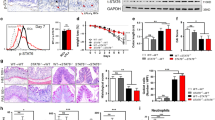
As we have previously identified that knocking down BVES with either siRNA or dominant-negative approaches led to epithelial junctional compromise in vitro,15 we hypothesized that the Bves−/− colon might also have abnormalities in tight junction-associated phenotypes such as permeability. While ultrastructural analysis of tight junctional complexes in Bves−/− colonic epithelium was unremarkable, measurements of intercellular lateral membrane boundaries demonstrated dilated intercellular spaces in Bves−/− colons as compared with WT (Fig. 2a). Consistent with this observation, Bves−/− ex vivo cultures maintained in Ussing chambers demonstrated increased paracellular movement of FITC–Dextran complexes after 3 h, indicating barrier incompetency (Fig. 2b). Finally, 3D Bves−/− colon organoids displayed reduced TER when grown on transwells as a 2D monolayer (Fig. 2c). Together, these data indicate that the epithelial barrier is compromised in Bves−/− mice.
Bves−/− colons display increased permeability and altered tight junction composition. a Intercellular spaces quantified by electron microscopy (Bves−/− vs. WT 44.0 ± 3.3 nm vs. 29.6 ± 1.3 nm, ***P < 0.0001, n = 100–105 images from three mice). Images taken at ×42,000. White arrows label tight junctions and black arrowheads delineate intercellular spaces on the basolateral side. b Amount of penetrated FITC–dextran was quantified via Ussing chamber ex vivo permeability studies (Bves−/− vs. WT, 0.020 ± 0.003 vs. 0.003 ± 0.001 μg/mL, *P < 0.05, n = 4 in each group). c Colonoids were isolated and grown as monolayers on transwells, and transepithelial resistance (TER) was measured daily by Ohm meter. TER differences were quantified on each day, but only day 4 and day 5 exhibited significant differences: Day 4 (Bves−/− vs. WT, 371.7 ± 4.8 vs. 522.2 ± 5.7 ohms, **P < 0.01, n = 6) and day 5 (326.5 ± 6.8 vs. 447.7 ± 8.2 ohms, **P < 0.01, n = 6). d Claudin 7 immunoblot from colonic crypts (left) and quantification (right; Bves−/− vs. WT, 70% reduction, *P < 0.05, n = 4). e Immunofluorescence of the colonic epithelium shows redistribution of pMLC, although pMLC and MLC immunoblot were nonsignificant (n = 4). Scale bars are shown in the bottom right and are 100 μM. f Tight junction mRNA expression of Bves−/− and WT crypt isolates. Claudin2 (Bves−/− vs. WT, 1.8-fold increase, *P < 0.05, n = 7–8); JAM-A (Bves−/− vs. WT, 1.6-fold increase, *P < 0.05, n = 7–8); Zo-1 (Bves−/− vs. WT, 1.4-fold increase, *P < 0.01, n = 7–8); Claudin1, Claudin3, and Claudin11 (P = ns, n = 7–8)
We next examined tight junction composition of the colonic crypts using a candidate gene approach. Immunoblot for Claudin 7 (Fig. 2d) showed a significant decrease in protein levels. However, there was a trend toward increase in ZO-1 protein expression. We then performed qPCR for several other tight junction proteins (Fig. 2f). There was a significant increase in Claudin2, JAM-A, and Zo-1 mRNA expression. There was no change in mRNA expression in Claudin1, Claudin3, or Claudin11. Given that BVES has previously been shown to regulate RhoA signaling in the LIM2405 CRC cell line,15 we immunoblotted for phosophomyosin light chain 2 (pMLC) and myosin light chain 2 (MLC). While the immunoblot for pMLC and total MLC did not show a difference between WT and Bves−/− colonic crypt isolates, immunofluorescence of the epithelium shows an increase in pMLC staining on the luminal side of the crypt (Fig. 2e). We then compared adherens junctions proteins E-cadherin and β-catenin expression in colonic crypts by immunoblot and/or immunofluorescence and saw no difference in levels or localization (Supplementary Figure 2A-B). Finally, immunoblot showed a significant increase in myosin 2C expression, while immunofluorescence for phalloidin, myosin 2B, and myosin 2C showed no difference in localization (Supplementary Figure 3A-C). Thus, BVES deletion leads to dysregulation of several tight junction-associated proteins in the colon, with the net effect being increased permeability.
Bves −/− mice display increased lymphoid aggregates at baseline
Since Bves−/− mice exhibit increased permeability, we hypothesized that they would have alterations in immune cells within the colon. We first observed that there were increased lymphoid aggregates in the colonic mucosa in Bves−/− mice (Fig. 3a) with an increase in fraction of T cells and myeloid cells (Fig. 3b,c) and no detectable difference in B cells within these lymphoid aggregates (Fig. 3d).
Bves−/− colons have increased lymphoid aggregates. Colons were isolated and swiss-rolled. For each analysis images are on left and quantification on right. a Total number of lymphoid aggregates per colon (Bves−/− vs. WT, 2.5 vs. 1.2, *P < 0.05, n = 7–10). Scale bar in bottom left corner and is 500 μM. b CD3 immunohistochemistry and CD3+ cells/lymphoid aggregate (Bves−/− vs. WT, 62.4 vs. 39.7, *P < 0.05, n = 5). c Neutrophil immunofluorescent staining (Ly6B.2) and Ly6B.2+ cells/lymphoid aggregate (DAPI = blue, E-cadherin = green, and Ly6B.2 = red; Bves−/− vs. WT, 2.7 vs. 0.5%, *P < 0.05, n = 4–5). d B-cell immunofluorescence (B220) and B220+ cells/lymphoid aggregate (DAPI = blue, E-cadherin = green, and B220 = red; Bves−/− vs. WT, 48.6 vs. 46.7%, P = 0.56). For b–d, scale bars are in the bottom left corner and represent 100 μm
BVES maintains colonic epithelial integrity after C. rodentium colitis
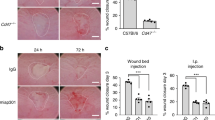
In order to analyze whether the aforementioned morphometric changes (i.e., crypt hyperplasia), increased permeability, and increased lymphoid aggregate population increased the susceptibility of Bves−/− mice to develop experimental colitis, we inoculated mice with C. rodentium, which is a widely used model to induce colonic epithelial injury and is useful in interrogating intestinal repair responses. Both WT and Bves−/− mice lost weight after C. rodentium inoculation; however, by day 3 post inoculation, Bves−/− mice trended toward increased weight loss from day 4 to day 14 (Fig. 4a). At necropsy, the colon weight as a percent of body weight as well as colon weight/colon length was significantly greater in the Bves−/− mice (Fig. 4b,c). Histological review of H&E-stained Swiss-rolled colons (Fig. 4d) by a gastrointestinal pathologist examining metrics of epithelial ulceration, inflammatory infiltrate, submucosal edema, and impaired regeneration revealed worse injury in Bves−/− colons (Fig. 4e). Bves−/− mice also demonstrated increased colonization of the colonic mucosa (Fig. 4f) and mesenteric lymph nodes (Fig. 4g) after C. rodentium infection.
Bves−/− mice demonstrated augmented colitis after infection with Citrobacter rodentium. WT and Bves−/− mice were infected with C. rodentium and killed 14 days after inoculation. a Body weight measured daily for 14 days after inoculation shown as a percentage of starting weight on each day (Bves−/− vs. WT, P = ns, n = 8–14). b Ratio of colon weight to body weight at the time of killing (Bves−/− C. rod vs. WT C. rod, 1.73 ± 0.06 vs. 1.40 ± 0.08, ***P < 0.001, n = 8–14). c Ratio of colon weight to colon length at the time of killing (Bves−/− vs. WT, 0.04 ± 0.001 vs. 0.03 ± 0.002, ***P < 0.001, n = 8–14). d Representative H&E-stained sections of infected WT and Bves−/− mice. Scale bars are pictured in the lower left hand corner. They represent 500 μM for ×5 images on left, while they represent 100 μM for the ×20 images on the right. e Injury scores (Bves−/− vs. WT, 6.9 ± 1.3 vs. 11.0 ± 1.1% **P < 0.01, n = 11–14). Histological injury score using a multipoint scale as described in the Materials and methods section. f Bacterial colonization of colon with C. rodentium quantified as log (CFU/g colon; Bves−/− vs. WT, 7.4 ± 0.4 vs. 6.8 ± 0.2, *P < 0.05, n = 8–13). g Bacterial colonization of mesenteric lymph nodes (mLNs) calculated as CFU/g mLN (Bves−/− vs. WT, 224 ± 99 vs. 773 ± 156 CFU/g, *P < 0.05, n = 6). h Colonocyte proliferation was calculated as a ratio of the number of pH3-positive-stained cells after treatment with C. rodentium divided by number of pH3-positive-stained cells at baseline (Bves−/− Injury/Baseline vs. WT Injury/Baseline, 4.5 ± 0.4 vs. 8.1 ± 3.7, **P < 0.01, n = 11–13). i mRNA expression of stem cell markers from colon epithelial isolates at baseline quantified by qPCR: cMyc (Bves−/− vs. WT, 1.8 ± 0.2 vs. 1.0 ± 0.2-fold change, *P < 0.05, n = 7–8), Lgr5 (Bves−/− vs. WT, 1.5 ± 0.3 vs. 1.0 ± 0.2 P = 0.16, n = 8), and Lrig1 (Bves−/− vs. WT, 2.1 ± 0.5 vs. 1.0 ± 0.4, P = 0.09, n = 7–8). j After C. rodentium infection, expression of stem cell markers relative to baseline was quantified by qPCR. cMyc (Bves−/− vs. WT, 0.1 ± 0.02 vs. 7.7 ± 0.8-fold change relative to baseline, **P < 0.01), Lgr5 (Bves−/− vs. WT, 0.004 ± 0.0005 vs. 5.8 ± 0.9-fold change relative to baseline, **P < 0.01), and Lrig1 (Bves−/− vs. WT, 75 ± 20 vs. 212 ± 40-fold change relative to baseline, P = 0.42)
One consequence of increased bacterial colonization may be that WT mice have a greater proliferative response than Bves−/− colons in response to C. rodentium. In other words, the increase in proliferation from pre-injury to post injury is greater in WT mice (Fig. 4h). This is important because increased epithelial proliferation must occur to efficiently reconstitute epithelial barrier properties. We hypothesized that an increased stem cell compartment could explain this change in proliferation, as an increased stem cell number would result in an increased number of proliferating cells. As such, we compared mRNA expression of stem cell markers from colon epithelial isolates both before and after C. rodentium infection. At baseline, cMyc was significantly upregulated in Bves−/− colon epithelium, while there was a trend toward an increase in stem cell markers Lgr5 and Lrig1 (Fig. 4i). After C. rodentium infection, cMyc and Lgr5 were both upregulated in WT while they were both downregulated in Bves−/− mice relative to baseline. Although not statistically significant, Lrig1 was upregulated after C. rodentium in both WT and Bves−/− mice but to a lesser degree in Bves−/− mice (Fig. 4j). Finally, we found no differences in apoptosis (Supplementary Figure 4) between cohorts. As such, BVES loss predisposes mice to increased bacterial colonization (of both the colon and mesenteric lymph nodes) and thus more severe colitis. Moreover, Bves−/− mice have impaired injury-repair programs, as evidenced by its relative hypoproliferative response and compromised stem cell compartment.
Bves −/− mice demonstrate altered immune responses at baseline and after C. rodentium infection
Since C. rodentium uncovered increased histologic injury and impaired injury-repair programs, we hypothesized that Bves−/− mice would also have alterations in immune response. Tissue-specific chemokine and cytokine production was evaluated in Bves−/− colons using Luminex profiling. At baseline, there was increased protein expression of CCL5, IL-5, IL-15, and IL-1α, while there was decreased IL-1β, IL-6, and IL-17 (Fig. 5a). After C. rodentium infection, CXCL1, IL-6, IL-7, G-CSF, and IL-17 were increased in Bves−/− mice after adjusting for expression of these analytes at baseline (i.e., before infection, Fig. 5b). Supplemental Table 1 summarizes the levels of all analytes after adjusting for baseline levels of expression. Of note, CXCL1 is chemotactic for polymorphonuclear cells; G-CSF stimulates survival, proliferation, and function of both neutrophils and neutrophil precursors; IL-6 is macrophage-derived and a pro-inflammatory marker; IL-7 is derived from dendritic cells and epithelial cells and stimulates proliferation of B cells, T cells, and NK cells. Finally, IL-17 is from Th17 cells and innate lymphoid cells, and most importantly is a well-established part of the pro-inflammatory response to C. rodentium.35 Specific levels of protein expression are included in Supplemental Table 2 as well as all of the other chemokines and cytokines that were measured. Collectively, these observations suggest that Bves−/− colons are primed for a robust inflammatory response when exposed to C. rodentium, and there is a significantly altered immune response post infection.
Bves−/− colons display dysregulated chemokine and cytokine expression by Luminex multi-analyte profiling. a Chemokines and cytokines that were significantly different between WT and Bves−/− colon at baseline: CCL5 (**P < 0.01, n = 7–8), IL-1⍺ (*P < 0.05, n = 6–8), IL-5 (**P < 0.01, n = 3–7), IL-1β (*P < 0.05), IL-15 (*P < 0.05, n = 5–6), IL-6 (*P < 0.05, n = 5–8), and IL-17 (*P < 0.05, n = 6–8). b Chemokines and cytokines that are significantly different between WT and Bves−/− colons after C. rodentium: CXCL1 (*P < 0.05, n = 7), IL-6 (**P < 0.01, n = 6–7), IL-7 (***P < 0.001, n = 6–7), G-CSF (***P < 0.001, n = 7), and IL-17 (**P < 0.01, n = 6)
Bves −/− mice exhibit upregulated IL-22-mediated mucosal host defenses after C. rodentium
We next examined expression of IL-22, which is known to be upregulated after C. rodentium infection. IL-22-deficient mice exhibit more severe colitis after C. rodentium and thus is thought to mediate the host’s initial resistance.36 In addition, IL-22 is significantly upregulated in patients with CD and UC.37 Il22 mRNA expression in whole colon after C. rodentium infection is increased in Bves−/− mice (Fig. 6a). There is also significantly increased mRNA expression of IL-22 downstream targets, Reg3b, Reg3g, and Nos2 in colonic epithelial isolates (Fig. 6b). In addition, immunofluorescence for neutrophils reveals increased neutrophilic infiltration in Bves−/− mice after C. rodentium infection (Fig. 6c), consistent with increased CXCL1 expression observed in luminex profiling (Fig. 5b). The increase in neutrophils, Il22, and its downstream targets suggests that host defense mechanisms could be upregulated in response to neutrophilic infiltration more directly and increased bacterial colonization and attachment to the epithelium more indirectly.
Bves−/− mice exhibited upregulated IL-22-mediated mucosal host defenses after C. rodentium. a Il22 mRNA expression in whole colon after C. rodentium infection was quantified by qPCR (Bves−/− vs. WT, 2.3 ± 0.3 vs. 1.0 ± 0.2-fold change, *P < 0.05, n = 4). b IL-22 downstream targets are also quantified by qPCR in colonic epithelial isolates: Reg3b (Bves−/− vs. WT, 3.2 ± 0.4 vs. 1.0 ± 0.3-fold change, **P < 0.01, n = 5), Reg3g (Bves−/− vs. WT, 2.8 ± 0.4 vs. 1.0 ± 0.2-fold change, **P < 0.01, n = 5), and Nos2 (Bves−/− vs. WT, 1.8 ± 0.2 vs. 1.0 ± 0.2-fold change, *P < 0.05, n = 5). c Neutrophils were quantified by immunofluorescence for Ly6B.2 (Bves−/− vs. WT, 28.8 ± 4.2 vs. 9.8 ± 2.7 neutrophils per 10 crypts, **P < 0.01, n = 7). Images shown are at ×20. Scale bars are pictured in the lower right-hand corner. They represent 100 μm
Bves −/− mice have worse colonic injury after DSS
We next determined whether BVES-dependent injury responses observed in infectious colitis modeling were generalizable to other methods of injury by treating WT and Bves−/− mice with DSS to induce a chemical colitis. Using colonoscopy to evaluate for colitis progression Bves−/− mice treated for 5 days with 4% DSS appeared to have more severe injury in comparison to WT control animals (Fig. 7a). Histologic examination of colonic H&E staining and ulceration scores confirmed increased injury phenotypes in Bves−/− colons (Fig. 7b). There was no difference in Cxcl1 mRNA expression or neutrophilic infiltration in the acute DSS trial (Supplementary Figure 5C-D). Notably, similar to what we observed in infectious injury modeling, Bves−/− mice did not exhibit a post-injury hyperproliferative response. In fact, there was a decrease in proliferation when adjusting for number of proliferating cells before DSS (Fig. 7c).
Bves−/− mice demonstrated more severe colitis after DSS treatment. a WT (n = 10) and Bves−/− mice (n = 13) were treated with 4% DSS for 5 days and then sacrificed. Colonoscopy was performed prior to being sacrficed. Colonoscopy videos were scored using MEICS (Bves−/− vs. WT, 6.0 ± 0.6 vs. 3.3 ± 0.6, **P < 0.01, n = 10–13). b Acute injury scores quantified after sacrifice (Bves−/− vs. WT, 0.7 ± 0.1 vs. 0.4 ± 0.07, *P < 0.05, n = 6–7). Scale bars are pictured in the lower left hand corner. They represent 100 μM. c Left: proliferation was defined by pH3-positive-stained cells/crypt in the WT mice as compared with Bves−/− mice (Bves−/− vs. WT, 0.45 ± 0.08 vs. 0.53 ± 0.2, P = ns, n = 7). Right: proliferation was calculated as a ratio of the number of pH3-positive-stained cells after treatment with DSS divided by number of pH3-positive-stained cells at baseline (Bves−/− Injury/Baseline vs. WT Injury/Baseline, 0.3 ± 0.05 vs. 1.0 ± 0.2, **P < 0.01, n = 6–7). d Injury-repair model. Mice were treated with 4% DSS for 5 days and then allowed to recover for 5 days. The mice were then killed. There was significantly greater histologic injury in the Bves−/− mice (0.6 ± 0.08 vs. 0.3 ± 0.03, **P < 0.01, n = 4–5). Scale bars are pictured in the lower left hand corner. They represent 100 μM. e Left: increased proliferation defined by pH3-positive-stained cells/crypt in the WT mice as compared with Bves−/− mice (Bves−/− vs. WT, 0.65 ± 0.1 vs. 1.3 ± 0.2, *P < 0.05, n = 4–5). Right: there was also increased proliferation when compared with mice not treated with DSS (i.e., when compared with baseline proliferation; Bves−/− Injury/Baseline vs. WT Injury/Baseline, 0.4 ± 0.1 vs. 2.2 ± 0.4, *P < 0.05, n = 4–5). f Survival curves from mice treated with three cycles of 4% DSS for 5 days and 10 days of recovery (Bves−/− vs. WT, 62.5 vs. 100%). Survival curves were compared using Log-rank (Mantel–Cox) test. *P < 0.05, n = 8–9
We next tested for repair process deficiencies by treating WT and Bves−/− mice with 4% DSS for 5 days and then allowed them to recover for 5 days (DSS replaced with water). While there was similar weight loss in both genotypes in both the acute and injury-repair DSS trials (Supplementary Figure 5A-B), histologic analysis determined that there was greater injury in Bves−/− mice (Fig. 7d) and that proliferation was decreased in Bves−/− mice as compared with WT after the recovery period both before and after adjusting for pre-DSS proliferation (Fig. 7e).
Finally, to model response to chronic epithelial injury, we treated WT and Bves−/− mice with 4% DSS and then allowed them to recover for 10 days, three times consecutively. After three cycles of 4% DSS, 100% of WT mice had survived, but only 62.5% of Bves−/− mice survived (Fig. 7f). While there was not a significant difference in histologic ulceration score or proliferation in the surviving mice, this could potentially be due to loss of more moribund mice in the Bves−/− group, preventing histologic examination of the sicker mice. Thus, consistent with our results in the C. rodentium model of colitis, DSS treatment of Bves−/− mice yields greater histologic injury in acute and injury-repair modeling and increased mortality rates in a chronic model.
BVES limits EPEC infection in human cell lines
To dissect the mechanism by which Bves−/− mice were more sensitive to colitogenic insult, we first studied the effect of EPEC, a C. rodentium human analog, on isolated epithelial cell cultures. We infected Caco2 cells, a polarized CRC-derived epithelial cell line, with EPEC as it has a similar mechanism of pathogenesis as C. rodentium, namely formation of attaching and effacing lesions and resultant epithelial damage. After knocking down BVES (Fig. 8a) via an siRNA approach, 3 h EPEC infection resulted in increased CXCL8 expression (Fig. 8b), which is known to be induced by EPEC infection and is an indicator of bacterial invasion.38 There is also increased attachment of EPEC as evidenced by actin stalks (Fig. 8c). These observations were replicated in HEK293T cells (Fig. 8d). We observed a consistent phenotype with increased expression of CXCL8 (Fig. 8e) and increased attaching/effacing lesions per cell (Fig. 8f) after knocking down BVES. Moreover, BVES-transfected HEK293T cells (Supplemental Fig. 6A) were then infected with EPEC for 3 h. In these experiments, we observed decreased CXCL8 expression (Supplemental Fig. 6B) as well as a decreased number of attaching/effacing lesions by actin immunofluorescence in cells overexpressing BVES (Supplemental Fig. 6C). These findings suggest that the pathogenicity of EPEC and hence C. rodentium can be limited by BVES expression.
BVES influences pathogenicity of EPEC in Caco2 and HEK cells. a BVES expression after siRNA knockdown in Caco2 cells (siBVES vs. scr, 0.06 ± 0.01 vs. 1.0 ± 0.16, **P < 0.01). b CXCL8 mRNA expression quantified after 3 h EPEC infection in Caco2 cells (siBVES vs. scr, 17 ± 4.5-fold vs. 4.1 ± 0.4-fold, **P < 0.01, two experiments with three replicates each time). c Number of actin stalks per Caco2 cell after 3 h EPEC infection (siBVES vs. scr, 9.1 ± 0.9 vs. 4.0 ± 0.6, ***P < 0.001, two experiments with three replicates each time). Green dots represent individual actin stalks. Images are taken at ×80. d BVES knockdown by an siRNA approach in HEK293T cells (scr vs. siBVES, 1.0 ± 0.2 vs. 0.08 ± 0.01, *P < 0.05). e CXCL8 expression mRNA expression quantified after 3 h EPEC infection in HEK293T cells (scr vs. siBVES, 2.0 ± 0.2-fold vs. 8.1 ± 1.5-fold, *P < 0.05, two experiments with three replicates each time). f Number of actin stalks per HEK293T cell quantified after 3 h EPEC infection (scr vs. siBVES, 8.4 ± 0.5 vs. 9.9 ± 0.5, *P < 0.05)
BVES is underexpressed in moderate/severe UC
On the basis of the above findings, we next examined clinical samples to determine whether BVES levels were altered in UC. Clinical subjects were recruited from control and IBD subjects undergoing colonoscopy for evaluation of UC or non-UC indications at Vanderbilt University. Mucosal biopsies were obtained from control subjects and UC patients from areas of active disease, which was confirmed based on histological analysis.28 RNA was extracted, qRT-PCR performed, and delta–delta Ct calculations between normal and disease samples indicated a 2.3-fold reduction of BVES mRNA levels in moderate/severe UC samples (Fig. 9a). The patient characteristics, including age, gender, BMI, and current medications, are shown in Fig. 9b. These observations are consistent with our experimental data and suggest that BVES may also contribute to epithelial integrity in IBD.
BVES is underexpressed in moderate to severe ulcerative colitis. BVES mRNA levels were determined using qRT-PCR on UC biopsy specimens. a UC status determined by histologic status. aP < 0.05, bP < 0.01, cP < 0.001 vs. control. *This patient was started on therapy for presumed UC but with normal endoscopic evaluation was felt to have irritable bowel syndrome. Age and body mass index statistics were assessed by the Kruskal–Wallis test followed by the Mann–Whitney test. b BVES mRNA levels expressed as negative ddCt in normal, quiescent/mild UC, and moderate/severe UC. BVES expression quantified between normal and moderate/severe UC (*P < 0.05, n = 7–8, Kruskal–Wallis) and between quiescent/mild and moderate/severe UC (*P < 0.05, n = 7–16, Kruskal–Wallis)
Discussion
In this study, we investigated the role of BVES in colonic homeostasis and in response to colon injury. At baseline, Bves−/− mice have a number of subtle colonic phenotypes, which in aggregate we postulated might result in altered injury response when challenged. In every experimental model we tested, Bves−/− mice had exacerbated colitis, and in the C. rodentium model there was increased bacterial translocation. EPEC infection of both Caco2 and HEK293T BVES knockdown cell lines increased the number of attaching/effacing lesions. Conversely, fewer attaching/effacing lesions occurred when BVES expression was increased in these lines, suggesting that BVES limited colonization. Lastly, BVES expression was reduced in UC samples, indicating relevance to human disease and a potential protective role for BVES in human disease.
We propose that colitis may be more severe in the absence of BVES because of compromised barrier function. Redistribution of phosphomyosin light chain (pMLC) to the luminal epithelial surface could potentially alter apical web contractile properties and thus increase permeability.39 Interestingly, we had previously observed that when BVES function was impaired in LIM2405 cells pMLC expression increased.15 In addition, the downregulation of Claudin-7 and concurrent increase in Claudin 2 could contribute to increased permeability and susceptibility to both infectious and chemical colitis.40,41 Further, we observed increased expression of ZO-142 and JAM-A,11 which function to preserve epithelial barrier function. These increases could represent compensatory changes arising with BVES loss and potentially limit significant inflammation at baseline. However, these barrier defects may compromise the epithelium, priming for increased C. rodentium colonization and susceptibility to chemical colitis.
C. rodentium infection results in reproducible colonic inflammation and injury with fewer systemic effects in comparison to other models of infectious colitis.43,44 Two pathognomonic features of C. rodentium infection include induction of actin cytoskeletal reorganization causing attaching/effacing lesions in the colonic epithelium as well as development of crypt hyperplasia, which is triggered by the loss of epithelial barrier integrity and the transit of bacteria into the lamina propria.45 Knocking down BVES in both HEK293T and Caco2 cells increased the numbers of attaching/effacing lesions and increased Cxcl8 expression, indicating more severe infection. One possible explanation for increased attaching/effacing lesions is that the loss of BVES and subsequent loss of cell-cell adhesion increases possible binding sites for EPEC and C. rodentium. C. rodentium infection also induces robust Th1 and Th17 immune responses.46,47 After C. rodentium inoculation, Bves−/− mice demonstrated increased IL-17 protein expression and increased bacterial colonization and seeding of mesenteric lymph nodes, indicative of a more systemic infection. Moreover, increased Il22, Reg3b, Reg3g, and Nos2 expression strongly suggests that host mucosal defenses were upregulated in response to more severe disease. As such, loss of BVES increases the susceptibility of colonic epithelial cells to attaching/effacing bacteria and upregulates immune responses.
It was unexpected that Bves−/− mice would have a hypoproliferative response after injury, especially in the setting of potentiated proliferation at baseline. While we don’t have a unifying explanation for this phenotype at this time, one hypothesis is that injury resulted in a relative depletion of stem cells in Bves−/− mice resulting in reduced inflow into the transit amplifying cell compartment. This suggests that a role for BVES in stem cell preservation after injury. We speculate that increased stem cell activity at baseline places these populations at increased risk when injured.
Future experiments will further elucidate the mechanisms by which the stem cell compartment is compromised after chemical or infectious injury. We also plan to analyze the interaction between BVES and other tight junction proteins, particularly Claudin-7 and Claudin 2, which are dysregulated in Bves−/− mice. Additionally, our group has reported that BVES interacts with a subunit of PP2A,21 which has previously been implicated in acetaldehyde-induced disruption of tight junctions in Caco2 monolayers.48 We plan to investigate the functional relevance of the BVES-PP2A interaction after injury C. rodentium and EPEC.
In conclusion, our findings demonstrate that BVES contributes to multiple aspects of intestinal homeostasis and colonic injury responses. Defects in other TJ constituents have compromised epithelial barrier function and lead to increased paracellular permeability,11,12,40,41 similar to the phenotypes we observed with BVES loss. For example, loss of tight junction-associated proteins JAM-A and NMIIA resulted in baseline inflammation and increased susceptibility to chemical colitis with DSS treatment.11,12 Furthermore, TJ dysfunction is implicated in the pathogenesis of IBD. Prior studies have demonstrated that altered permeability due to tight junction disruption is associated with both increases in bacterial colonization and bacterial load, as well as hyperactive mucosal immune responses.49,50 In these experiments, we identify a critical role for a tight junctional protein, BVES, in preserving colonic epithelial integrity under physiologic stress induced by both chemical and infectious agents. Bves−/− mice display increased permeability, have altered tight junction composition, have increased lymphoid aggregates, have worse colitis after DSS treatment, and have increased bacterial colonization and histologic injury after infection with C. rodentium. Finally, loss of BVES expression in Caco2 and HEK293T cells leads to increased attaching/effacing lesions after infection with EPEC, while these lesions are decreased with augmented BVES expression. These findings, combined with decreased expression of BVES in patients with UC, suggest that alterations in BVES may be relevant to understanding the mechanisms that underlie epithelial barrier dysfunction in colitis.
References
Shivashankar, R. et al. Incidence and prevalence of Crohn’s disease and ulcerative colitis in Olmsted county, Minnesota from 1970 through 2010. Clin. Gastroenterol. Hepatol. 15, 857–863 (2016).
Baumgart, D. & Sandborn, W. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet 369, 1641–1657 (2007).
Maloy, K. J. & Powrie, F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306 (2011).
Sturm, A. & Dignass, A. U. Epithelial restitution and wound healing in inflammatory bowel disease. World J. Gastroenterol. 14, 348–353 (2008).
Iizuka, M. & Konno, S. Wound healing of intestinal epithelial cells. World J. Gastroenterol. 17, 2161–2171 (2011).
Söderholm, J., Olaison, G. & Peterson, K. Augmented increase in tight junction permeability by luminal stimuli in the non-inflamed ileum of Crohn’s disease. Gut 50, 307–313 (2002).
Shen, L., Su, L. & Turner, J. R. Mechanisms and functional implications of intestinal barrier defects. Dig. Dis. 27, 443–449 (2009).
Balda, M. S. et al. Structure, regulation, and pathophysiology of tight junctions in the gastrointestinal tract. Yale J. Biol. Med. 65, 725–735 (1992).
Balda, M. S. & Matter, K. Tight junctions. J. Cell Sci. 111, 541–547 (1998). Pt 5.
Turner, J. R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 9, 799–809 (2009).
Laukoetter, M. G. et al. JAM-A regulates permeability and inflammation in the intestine in vivo. J. Exp. Med. 13, 3067–3076 (2007).
Naydenov, N. G. et al. Nonmuscle MyosinIIA regulates intestinal epithelial barrier in vivo and plays a protective role during experimental colitis. Sci. Rep. 6, 24161 (2016).
Reese, D. et al. Bves: a novel gene expressed during coronary blood vessel development. Dev. Biol. 209, 159–171 (1999).
Andrée, B. et al. Isolation and characterization of the novel popeye gene family expressed in skeletal muscle and heart. Dev. Biol. 223, 371–382 (2000).
Williams, C. S. et al. BVES regulates EMT in human corneal and colon cancer cells and is silenced via promoter methylation in human colorectal carcinoma. J. Clin. Invest. 121, 4056–4069 (2011).
Andrée, B. et al. Mouse Pop1 is required for muscle regeneration in adult skeletal muscle. Mol. Cell Biol. 22, 1504–1512 (2002).
Kawaguchi, M. et al. Identification of a novel intracellular interaction domain essential for Bves function. PLoS ONE 3, e2261 (2008).
Vasavada, T. K., DiAngelo, J. R. & Duncan, M. K. Developmental expression of Pop1/Bves. J. Histochem. Cytochem. 52, 371–377 (2004).
Jayagopal, A. et al. Tight junction-associated signaling pathways modulate cell proliferation in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 52, 588–593 (2011).
Reddy, V. K. et al. BVES regulates intestinal stem cell programs and intestinal crypt viability after radiation. Stem Cells 34, 1626–1636 (2016).
Parang, B. et al. BVES regulates c-Myc stability via PP2A and suppresses colitis-associated tumourigenesis. Gut 66, 852–862 (2017).
Madara, J. L. Increases in guinea pig small intestinal transepithelial resistance induced by osmotic loads are accompanied by rapid alterations in absorptive-cell tight-junction structure. J. Cell Biol. 97, 125–136 (1983).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011).
Williams, C. S. et al. MTG16 contributes to colonic epithelial integrity in experimental colitis. Gut 62, 1446–1455 (2013).
Singh, K. et al. The apolipoprotein E-mimetic peptide COG112 inhibits the inflammatory response to Citrobacter rodentium in colonic epithelial cells by preventing NF-kappaB activation. J. Biol. Chem. 283, 16752–16761 (2008).
Singh, K. et al. The apolipoprotein E-mimetic peptide COG112 inhibits NF-kappaB signaling, proinflammatory cytokine expression, and disease activity in murine models of colitis. J. Biol. Chem. 286, 3839–3850 (2011).
Gobert, A. P. et al. Protective role of arginase in a mouse model of colitis. J. Immunol. 173, 2109–2117 (2004).
Coburn, L. A. et al. High-throughput multi-analyte Luminex profiling implicates eotaxin-1 in ulcerative colitis. PLoS One 8, e82300 (2013).
Barrett, C. W. et al. Tumor suppressor function of the plasma glutathione peroxidase Gpx3 in colitis-associated carcinoma. Cancer Res. 73, 1245–1255 (2012).
Barrett, C. W. et al. MTGR1 is required for tumorigenesis in the murine AOM/DSS colitis-associated carcinoma model. Cancer Res. 71, 1302–1312 (2011).
Becker, C., Fantini, M. C. & Neurath, M. F. High resolution colonoscopy in live mice. Nat. Protoc. 6, 2900–2904 (2006).
Mahler, M. et al. Differential susceptibility of inbred mouse strains to dextran sodium sulfate-induced colitis. Am. J. Physiol. 274, G544–G551 (1998).
Yan, C. et al. Epithelial to mesenchymal transition in human skin wound healing is induced by tumor necrosis factor-α through bone morphogenic protein-2. Am. J. Pathol. 176, 2247–2258 (2010).
Bataille, F. et al. Evidence for a role of epithelial mesenchymal transition during pathogenesis of fistulae in Crohn’s disease. Inflamm. Bowel Dis. 14, 1514–1527 (2008).
Harrington, L. E. et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6, 1123–1132 (2005).
Zheng, Y. et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 (2008).
Andoh, A. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology 129, 969–984 (2005).
Meraz, I. et al. Epithelial cells secrete interleukin-8 in response to adhesion and invasion of diffusely adhering Escherichia coli&& lacking Afa/DrGenes. Micobiol. Immunol. 50, 159–169 (2006).
Citalán-Madrid, A. et al. Cortactin deficiency causes increased RhoA/ROCK1-dependent actomyosin contractility, intestinal epithelial barrier dysfunction, and disproprtionately severe DSS-induced colitis. Mucosal Immunol. 10, 1237–1247 (2017).
Tanaka, H. et al. Intestinal deletion of Claudin-7 enhances paracellular organic solute flux and initiates colonic inflammation in mice. Gut 64, 1529–1538 (2015).
Luettig, J. et al. Claudin-2 as a mediator of leaky gut barrier during intestinal inflammation. Tissue Barriers 3, e977176 (2014).
Groschwitz, K. R. et al. Intestinal barrier function: molecular regulation and disease pathogenesis. J. Allergy Clin. Immunol. 124, 3–22 (2009).
Borenshtein, D., McBee, M. E. & Schauer, D. B. Utility of the Citrobacter rodentium infection model in laboratory mice. Curr. Opin. Gastroenterol. 24, 32–37 (2008).
Eckmann, L. Animal models of inflammatory bowel disease: lessons from enteric infections. Ann. N. Y. Acad. Sci. 1072, 28–38 (2006).
Gibson, D. L. et al. Interleukin-11 reduces TLR4-induced colitis in TLR2-deficient mice and restores intestinal STAT3 signaling. Gastroenterology 139, 1277–1288 (2010).
Higgins, L. M. et al. Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect. Immun. 67, 3031–3039 (1999).
Mangan, P. R. et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature 441, 231–234 (2006).
Dungan, M. et al. Acetaldehyde disrupts tight junctions in Caco-2 cell monolayers by a protein phosphatase 2A-dependent mechanism. Am. J. Physiol. Gastrointest. Liver Physiol. 303, 1356–1364 (2012).
Darfeuille-Michaud, A. et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology 115, 1405–1413 (1998).
Macfarlane, G. T., Furrie, E. & Macfarlane, S. Bacterial milieu and mucosal bacteria in ulcerative colitis. Novartis. Found. Symp. 263, 57–64 (2004).
Acknowledgements
We thank members of the Williams and Wilson laboratories for thoughtful discussions about this research project. This work was supported by National Institutes of Health grants R01DK099204 and K08DK080221 to C.S.W., 5T32DK00767322 to Y.A.C., 1F30DK103498 and T32GM07347 to V.K.R., R01AT004821 to K.T.W., P50CA095103 to M.K.W., T32GM007347 to J.J.T., 1F30DK096718 to B.P., 1F31CA167920 to C.W.B., 5F32 DK108492 to S.P.S.; P30DK058404 (Vanderbilt Digestive Disease Research Center); American Cancer Society Research Scholar Grants 116552 to C.S.W.; Office of Medical Research, Department of Veterans Affairs (Merit Review Grants 1I01 BX001426 to C.S.W. and 2I01 BX001453 to K.T.W., and a Career Development Award 1IK2 BX002126 to L.A.C.); and Medical Research Council (MR/J010383/1) to T.B. Experiments/Data Analysis of TEM was performed in part through the use of the Vanderbilt Cell Imaging Shared Resource, supported by NIH grants CA68485, DK20593, DK58404, DK59637, and EY08126. We would like to thank the Vanderbilt TPSR core for staining, and the Vanderbilt Antibody and Protein Resource core for R-spondin. This publication was also supported in part by the National Cancer Institute Cancer Center Support Grant P30CA068485 and by CTSA award UL1TR000445 from the National Center for Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Choksi, Y.A., Reddy, V.K., Singh, K. et al. BVES is required for maintenance of colonic epithelial integrity in experimental colitis by modifying intestinal permeability. Mucosal Immunol 11, 1363–1374 (2018). https://doi.org/10.1038/s41385-018-0043-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-018-0043-2