Abstract
Inhaled pathogens including Pseudomonas aeruginosa initially encounter airway epithelial cells (AECs), which are poised to evoke cell-intrinsic innate defense, affecting second tier of hematopoietic cell-mediated immune reaction. However, it is largely unknown how pulmonary immune responses mediated by a variety of immune cells are coordinated. Here we show that Regnase-1, an endoribonuclease expressed in AECs and immune cells, plays an essential role in coordinating innate responses and adaptive immunity against P. aeruginosa infection. Intratracheal treatment of mice with heat-killed P. aeruginosa resulted in prolonged disappearance of Regnase-1 consistent with sustained expression of Regnase-1 target inflammatory genes, whereas the transcription factor NF-κB was only transiently activated. AEC-specific deletion of Regnase-1 not only augmented innate defenses against P. aeruginosa but also enhanced secretion of Pseudomonas-specific IgA and Th17 accumulation in the lung, culminating in conferring significant resistance against P. aeruginosa re-infection in vivo. Although Regnase-1 directly controls distinct sets of genes in each of AECs and T cells, degradation of Regnase-1 in both cell types is beneficial for maximizing acquired immune responses. Collectively, these results demonstrate that Regnase-1 orchestrates AEC-mediated and immune cell-mediated host defense against pulmonary bacterial infection.
Similar content being viewed by others
Introduction
The lower respiratory tract is constantly exposed to the air which contains microorganisms, some of which are potentially pathogenic. Failure to eliminate pathogens results in chronic respiratory infection or the development of pneumonia, both of which are leading causes of death in many countries. Among pathogens of the airway, Pseudomonas aeruginosa causes virulent acute and chronic infection, and is especially common in patients in intensive care or with compromised immune systems.1 In addition, P. aeruginosa often shows broad resistance against conventional antibiotic medications, and this characteristic leads to a high rate of treatment failure in clinical settings.2 Therefore, novel options for the treatment against P. aeruginosa infection are urgently required.
The respiratory immune reactions to pathogen infection are comprised of multiple tiered cellular responses, which function in a stepwise manner.3 The first tier of defense is mediated by innate immune cells that sense invasion of pathogens to mount innate responses and produce first-order cytokines.4 This is followed by second tier responses governed by lymphocytes inducing effector cytokines and producing pathogen-specific antibodies (Abs).4,5 In addition to hematopoietic cells, airway epithelial cells (AECs) play unique roles in the first line host defense at earliest stages of infection by acting as a physical barrier as well as mediating cell-intrinsic innate immunity.6,7 AECs secrete endogenous antibacterial agents such as lactoferrin, mucins, or surfactant proteins even under steady state.8 Furthermore, AECs sense invasion of pathogens via pattern-recognition receptors such as Toll-like receptors (TLRs) and secrete cytokines and chemokines to alert tissue-resident lymphocytes,3,6,9,10 although the importance of AECs in lymphocyte-mediated immune reactions has yet to be understood.
TLR signaling is known to induce genes involved in immune responses via transcription factors such as NF-κB and AP-1.9 Indeed, LPS treatment activated NF-κB in the lung.11 However, the activation was transient and returned to the basal level within 6 h.11 In contrast, LPS treatment induces prolonged inflammatory changes in the lung, which includes the upregulation of IL-6 and IL-12p40, and the upregulation sustains for more than 4 days.12 Thus, the mechanisms of sustaining inflammatory responses in the lung are yet to be understood.
In addition to transcriptional regulation, the activation of immune cells is controlled at post-transcriptional levels by a set of RNA-binding proteins, such as AU-rich element-binding proteins and stem-loop-binding proteins, including Roquin and Regnase-1.13,14,15,16,17 Among these, Regnase-1 (also known as MCPIP1) functions as an RNase essential for the regulation of immune cell activation.18,19,20 Regnase-1 degrades the mRNAs of inflammatory genes, such as Il6, Cxcl1, and Icos in innate and acquired immune cells by recognizing stem-loop structures in their mRNA 3′ untranslated regions (3′UTR).18,19,20 The expression of Regnase-1 is dynamically controlled in the course of immune reactions; in macrophages, TLR stimulation induces Regnase-1 degradation via IκB kinase-mediated phosphorylation.21 On the other hand, Regnase-1 is cleaved by a MALT1/paracaspase complexed with BCL10 in T cells following T cell receptor (TCR) ligation.19,22 Given that degradation of Regnase-1 facilitates immune-related gene expression, Regnase-1 functions as key switch to prevent overactivation of immune responses under steady-state or during pathogen infection. Although Regnase-1 negatively regulated IL-17-mediated gene expression in the lung,23 little is known about its function in non-immune cells such as epithelial cells or its association with pulmonary infection.
In the present study, we found that intratracheal treatment of mice with heat-killed P. aeruginosa resulted in sustained disappearance of Regnase-1 in the lung, in contrast to the transient activation of NF-κB. The expression of Regnase-1 target inflammatory genes was sustained following heat-killed P. aeruginosa treatment, and mice lacking Regnase-1 in AECs showed much higher resistance against re-infection of virulent P. aeruginosa. Interestingly, in addition to innate responses, Regnase-1 in AECs controls local secretion of P. aeruginosa-specific IgA and Th17 cell accumulation. Regnase-1 governs distinct but functionally overlapping set of genes depending on cell type (AECs or T cells). Collectively, P. aeruginosa infection-induced dynamic degradation of Regnase-1 plays a key role in various cell types including epithelial as well as immune cells to coordinate local innate and acquired immunity against pathogens.
Results
P. aeruginosa infection dynamically changes Regnase-1 expression in the lung
Pulmonary bacterial infection is known to activate NF-κB in the lung.7,24 When we treated mice with heat-killed P. aeruginosa intratracheally, phosphorylation of NF-κBp65 was induced 1 h after the treatment (Fig. 1a). Mild decrease of IκBα was also observed, indicating that the NF-κB transcriptional program was rapidly activated following P. aeruginosa treatment. In addition, the expression of Regnase-1 only modestly decreased 2 h after P. aeruginosa treatment (Fig. 1a). However, when we checked Regnase-1 expression at later time points, surprisingly, Regnase-1 disappeared 6 h after treatment, and remained diminished for more than 2 days (Fig. 1b). In contrast, phosphorylation of NF-κBp65, as well as change in the expression of IκBα had ceased by 6 h (Fig. 1b), suggesting that NF-κB activation rapidly ceased in the lung after being treated with heat-killed P. aeruginosa. The sustained disappearance of Regnase-1 was also observed in mice following intratracheal administration of LPS in the lung (Fig. 1c), and the disappearance was diminished in mice lacking MyD88, an adaptor molecule critical for TLR signaling (Fig. 1d). In contrast, deficiency of BCL10, an adaptor critical for MALT1 activation, resulted in temporal decrease of Regnase-1 in response to heat-killed P. aeruginosa administration in the lung, although the Regnase-1 protein levels were restored 24 and 48 h after the administration (Fig. 1e). These results indicate that TLR-MyD88 signaling induces degradation of Regnase-1 in the lung in response to P. aeruginosa, whereas MALT1 also contributes to the regulation of Regnase-1 protein levels in the lung, especially at the later time points.
P. aeruginosa infection dynamically changes Regnase-1 expression in the lung. a, b Immunoblot image for indicated proteins in the lung at early phase (a) and latter phase (b) after intratracheal administration of heat-killed P. aeruginosa (1.0 × 107 cfu) to WT C57BL/6J mice. c Immunoblot image for Regnase-1 protein levels in the lung of WT C57BL/6J mice after intratracheal LPS administration (60 μg). d, e Immunoblot image for indicated proteins in the lung after intratracheal administration of heat-killed P. aeruginosa (1.0 × 107 cfu) to MyD88–/– mice (d) and Bcl10–/– mice (e). f RT-qPCR analysis of the indicated genes in WT C57BL/6J mice at the time points indicated after intratracheal administration of heat-killed P. aeruginosa (1.0 × 107 cfu) or PBS. Data are shown as the relative abundance compared with the average of PBS-treated mice (n = 3 for each group). g Immunoblot image for Ptgs2 proteins in the lung after intratracheal administration of heat-killed P. aeruginosa (1.0 × 107 cfu) to WT C57BL/6J mice. h IL-6 (left panel) or IL-12p40 (right panel) concentration in the lysate of lung from WT C57BL/6J mice at indicated time points after intratracheal administration of heat-killed P. aeruginosa (1.0 × 107 cfu). N.S. no stimulation. Images are the representative of two independent experiments (a–e, g). Data are shown as mean ± s.d. Two-tailed Student’s t-test was used for statistical analyses (f, h).
Consistent with the TLR activation, the expression of Nfkbia, a well-known target of NF-κB, but not Regnase-1, was upregulated 6 h after P. aeruginosa stimulation, although the upregulation was not sustained for 48 h (Fig. 1f). On the other hand, the expression levels of mRNAs controlled by both NF-κB and Regnase-1, such as Ptgs2, Cxcl1, Il12b, and Il6 remained upregulated for 48 h (Fig. 1f). Furthermore, PTGS2, IL-12p40, and IL-6 proteins increased in the lung both at 6 and 48 h after P. aeruginosa stimulation (Fig. 1g, h). We also found that the Regnase-1 mRNA was upregulated both at 6 and 48 h, which is consistent with a previous report (Fig. 1f).21
These data suggest that after P. aeruginosa treatment in the lung, compared with NF-κB activation, degradation of Regnase-1 plays a large role in the regulation of inflammatory genes at late time points.
Loss of Regnase-1 in AECs confers resistance against pulmonary P. aeruginosa re-infection
Lung comprises a variety of different cell types, although AEC is one of major cell types. Since we found that Regnase-1 is indeed expressed in primary cultured AECs (mouse tracheal epithelial cells, MTEC) (Supplementary Fig. 1a), we examined if Regnase-1 governs barrier functions in AECs and contributes to the resistance against bacterial infection by generating mice lacking Regnase-1 in AECs. We crossed Nkx2.1-Cre mice, which express the Cre transgene in AECs,25 with mice carrying Regnase-1 floxed allele and knockout allele, and thereby generated AEC-specific Regnase-1-deficient mice (Nkx2.1-Cre+ Regnase-1flox/–, AEC-Reg1-cKO mice). We utilized a mT/mG double-fluorescent Cre reporter lineage tracing system to evaluate the successful deletion of Regnase-1 specifically at the AECs.26 In this system, cells expressing Cre replace td Tomato with GFP after Cre-mediated excision. First, histological examination revealed that the GFP expression was observed in AECs in Nkx2.1-Cre+ mT/mG mice (Supplementary Fig. 1b). Flow cytometry analysis revealed that GFP+ cells were solely observed in the CD45− non-hematopoietic cell population, and the expression of tdTomato was diminished in GFP+ cells (Supplementary Fig. 1c). RT-qPCR analysis revealed that the sorted GFP+ cells, but not control CD45+ cells, expressed Ccsp, Aqp5, and Sftpc, which are representative markers of AECs, type I or type II alveolar epithelial cells, respectively (Supplementary Fig. 1d). We further sorted the CD45− GFP+ cell fraction in AEC-Reg1-cKO mT/mG mice, and confirmed the successful deletion of Regnase-1 by RT-PCR (Supplementary Fig. 1e). These results clearly demonstrate that the deletion of Regnase-1 in the AECs was successful in AEC-Reg1-cKO mice.
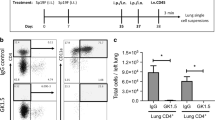
AEC-Reg1-cKO mice are viable, grow normally, and did not show any changes in the populations of T and B cells, macrophages, and neutrophils in the spleen (Supplementary Fig. 2a–c). Nevertheless, flow cytometry analysis revealed that there was significant increase of neutrophil population in the lungs of AEC-Reg1-cKO mice, compared with control mice (Fig. 2a, b). We confirmed that mRNA levels of Regnase-1 did not decrease in neutrophils from AEC-Reg1-cKO mice (Supplementary Fig. 3a). Treatment of wild-type (WT) mice with heat-killed P. aeruginosa for 48 h also increased neutrophil population in the lung, consistent with the degradation of Regnase-1 (Fig. 2c). These results suggest that Regnase-1 in AECs functions to restrict neutrophil accumulation under steady-state conditions.
Loss of Regnase-1 in AECs protects against pulmonary P. aeruginosa infection. a, c, e Neutrophil populations in untreated lung (a), heat-killed P. aeruginosa (1.0 × 108 cfu) treated lung (48 h after treatment) (c), or lung and BALF at 24 h after intratracheal 1.0 × 105 cfu PA103 infection (e) harvested from control and AEC-Reg1-cKO mice. Images are the representative of n > 3. b Total neutrophil count in the lung from untreated AEC-Reg1-cKO (n = 5) or control (n = 9) mice. d Hematoxylin and eosin (H&E) staining of lung samples from control or AEC-Reg1-cKO mice 24 h after intratracheal PA103 (1.0 × 105 cfu) infection (n = 3). A cluster of inflammatory cells (thick arrow) and infiltration of neutrophils into the alveolar space (thin arrow) are indicated. f Bacterial load in the lung from control (n = 7) or AEC-Reg1-cKO (n = 5) mice 24 h after intratracheal infection of 1.0 × 105 cfu PA103. g, h Body weight changes (n = 10) (g) and survival rate (control: n = 34, AEC-Reg1-cKO: n = 23) (h) after intratracheal 1.0 × 105 cfu PA103 infection. i Survival rate after the re-infection with 1.0 × 106 cfu PA103 on the day 29 of primary infection with 1.0 × 105 cfu PA103 (control: n = 15, AEC-Reg1-cKO: n = 7). Data are shown as mean ± s.d. Two-tailed Student’s t-test (b, g), Mann–Whitnely U test (f), or Wilcoxon test (h, i) was used for statistical analyses
When we intratracheally infected 1.0 × 105 cfu live PA103 strain-P. aeruginosa to AEC-Reg1-cKO or control mice, the accumulation of neutrophils into the alveolar space and interstitial tissue was more prominent in AEC-Reg1-cKO mice 24 h after the infection (Fig. 2d). In addition, neutrophil populations in the lung and bronchoaloveolar lavage fluid (BALF) increased in AEC-Reg1-cKO mice compared with controls (Fig. 2e). Interestingly, lack of Regnase-1 only in AECs enhanced competency to clear bacterial load in the lung 24 h after infection (Fig. 2f), and the rate of body weight loss was also significantly attenuated (Fig. 2g). Histologically, protraction of pulmonary inflammation was not observed in AEC-Reg1-cKO mice compared with control (Supplementary Fig. 3b). Consistent with the results above, AEC-Reg1-cKO mice tended to show resistance to initial P. aeruginosa infection compared with control mice (Fig. 2h), although the difference did not reach statistical significance.
Based on the results above, we hypothesized that Regnase-1 in AECs potentiated the first tier of defense, which can reduce the requirement of second tier mediated by lymphocyte activation. To examine the role of Regnase-1 in AECs in the resistance against recurrent bacterial infection which is cleared mainly by acquired immunity, we re-challenged the mice with P. aeruginosa PA103 with an increased bacterial dose (1.0 × 106 cfu) 4 weeks after the first challenge. In contrast to our hypothesis, AEC-Reg1-cKO mice showed strikingly higher resistance against re-infection compared with control mice (Fig. 2i). This result suggested the possibility that the loss of Regnase-1 in AECs contributes to the enhancement of acquired immunity in addition to natural and innate immune responses.
AEC Regnase-1 governs IgA secretion following P. aeruginosa infection
Since AEC-Reg1-cKO mice showed higher resistance against P. aeruginosa re-challenge, we tested if Ab responses are controlled by Regnase-1 in AECs. Interestingly, total IgA levels in BALF were much higher in P. aeruginosa-challenged AEC-Reg1-cKO mice than controls 22 days after infection (Fig. 3a). Furthermore, P. aeruginosa-specific IgA levels in BALF were also high in PA103-challenged AEC-Reg1-cKO mice (Fig. 3a). In sharp contrast, concentrations of total IgA and P. aeruginosa-specific IgA in the sera were comparable or reduced in AEC-Reg1-cKO compared with control mice after PA103 challenge (Fig. 3b), consistent with less bacterial load in the AEC-Reg1-cKO mice. These results suggest that Regnase-1 in AECs regulates local IgA production in the lung and/or secretion of IgA to the airway space.
AEC Regnase-1 governs IgA secretion following P. aeruginosa infection. a Total IgA (left panel) or P. aeruginosa-specific IgA (right panel) concentration of BALF from control or AEC-Reg1-cKO mice at 22 days after intratracheal PA103 (1.0 × 105 cfu) infection. Each dot represents one sample. b Total IgA (left panel) or P. aeruginosa-specific IgA (right panel) concentration of serum from control (n = 9) or AEC-Reg1-cKO mice (n = 6) at 22 days after intratracheal PA103 (1.0 × 105 cfu) infection. Each dot represents one sample. c Flow cytometry analysis for the plasma cell populations of the lung from control or AEC-Reg1-cKO mice on the day 22 after PA103 infection. Images are representative of n = 5. d The rate of IgA+ plasma cells among plasma cells in the lung from control or AEC-Reg1-cKO mice on the day 22 after PA103 infection. Image is representative of n = 5. e The rate of IgA+ plasma cells among lymphocytes in the lung from control or AEC-Reg1-cKO mice on the day 22 after PA103 infection. f IgA concentration of BALF or serum and secretory IgA concentrations from WT and Regnase-1–/– (Reg1–/–) mice evaluated by ELISA. g Secretory IgA concentrations from WT and Reg1–/– mice evaluated by ELISA. Mann–Whitney U test (a, b, g) and two-tailed Student’s t-test (e, f) was used for statistical analyses
To investigate the local production of IgA in the lung, we examined the population of IgA-producing plasma cells in the lung. Intratracheal infection of live P. aeruginosa PA103 to AEC-Reg1-cKO mice resulted in increase of total and IgA+ plasma cell populations compared with control mice on day 22 after infection (Fig. 3c–e). Collectively, Regnase-1 in AECs controls recruitment and/or generation of plasma cells specific to invading bacteria.
Next, we examined the possibility that Regnase-1 regulates secretion of IgA to the airway mucosa. We have previously shown that Regnase-1–/– mice (Reg1–/– mice) show severe pulmonary inflammation and high production of serum natural Abs including IgA.18 Consistently, total and IgA+ plasma cell populations increased greatly in the lung or mediastinal lymph node (mLN) of Reg1–/– mice (Supplementary Fig. 4a, b). Furthermore, the IgA concentration of BALF from Reg1–/– mice was elevated 15-fold or higher on average in control mice, and the increase in BALF was much higher even compared with the increase in serum (Fig. 3f). In addition, secretory IgA levels increased in Reg1–/– mice compared with control mice (Fig. 3g). Collectively, these data suggest that local IgA secretion as well as transcytosis to the BALF are enhanced in Reg1–/– mice.
Next, we evaluated if Regnase-1 alters commensal bacterial load in the lung, which can secondarily affect IgA secretion. qPCR analysis revealed that there was no statistical difference in the amounts of total as well as the filum-specific bacteria between control and Reg1–/– mice under quiescent conditions (Supplementary Fig. 4c), indicating that the increase of IgA secretion under Regnase-1 deficiency is not due to the alteration in commensal bacteria.
Identification of genes regulated by Regnase-1 in AECs
Next, we explored the mechanism how Regnase-1 in AECs controls lung immunity by performing transcriptome analysis using mRNAs prepared from WT and Reg1–/– MTEC. Among the genes significantly upregulated under Regnase-1 deficiency, we identified molecules with anti-microbial function (Fig. 4a). These genes included endogenous antimicrobials (e.g., Muc5b, Ltf, Lcn2), contributing to the mucosal barrier function. Interestingly, Pigr, a gene critical for the transcytosis of IgA in AECs,27 was found to be negatively regulated by Regnase-1 (Fig. 4b and Supplementary Fig. 5). Additionally, Regnase-1 expressed in AECs controls various chemokines and cytokines, including Csf3, Cxcl5, Ccl20, and Ccl28.28 CSF3 and CXCL5 contribute to the proliferation of neutrophils, CCL20 attracts Th17,29 and CCL28 induce the chemotaxis of plasma cells to the mucosal area,28 respectively. The upregulation of these genes in Reg1–/– MTEC was confirmed by RT-qPCR (Supplementary Fig. 5).
Genes associated with natural immunity and IgA secretion are upregulated in AECs lacking Regnase-1. a Scatterplot of gene expression profiles evaluated by RNA-sequencing for MTEC from WT or Reg1–/– mice. The yellow and blue lines represent 10-fold and 3-fold induction, respectively. P-values were calculated by using DEseq2. b Summary for upregulated genes in MTEC derived from Reg1–/– mice. Genes with RPKM <10 in WT-derived MTEC were excluded, and genes were ranked according to the ratio of gene expression levels between Reg1–/–-derived and WT-derived MTEC. c RT-qPCR analysis of the indicated genes in WT or Regnase-1 knockout (KO) Calu-3 cell line (n = 3 for each genotype). Data are shown as mean ± s.d. Two-tailed Student’s t-test (c) was used for statistical analyses
Next, we addressed whether this upregulation can be observed in human cells. For this purpose, we generated a human Reg1–/– AEC line (Calu-3) by using the CRISPR-Cas9 system (Supplementary Fig. 6a, b). Regnase-1-deficient Calu-3 showed increased expression of genes such as LCN2, PIGR, CXCL5, CCL28, and MUC5B (Fig. 4c). Reconstitution of Regnase-1 by a doxycycline-inducible system significantly suppressed these genes, indicating the roles of Regnase-1 in AECs are conserved between mouse and human (Supplementary Fig. 6c, d).
Loss of Regnase-1 enhances transcytosis and production of endogenous antibiotics
We next examined if the genes upregulated in MTEC are also suppressed by Regnase-1 in the lung tissue. Immunoblot analysis revealed that pIgR, Sftpd, and Muc5b proteins were much more abundantly expressed in Reg1–/– lung compared with controls (Fig. 5a), in concordance with the mRNA expression levels in vivo (Fig. 5b).
Loss of Regnase-1 enhances transcytosis and production of endogenous antibiotics. a Immunoblot for indicated protein levels in the lung of WT or Reg1–/– mice. Representative image of n = 3 for each genotype. b RT-qPCR analysis of the indicated genes in the lung from WT or Reg1–/– mice (n = 6 for each genotype). c Representative images of anti-IgA immunohistochemistry of lung samples from WT (n = 2) or Reg1–/– (n = 3) mice. d RT-qPCR analysis of the indicated genes in the lung from control or AEC-Reg1-cKO mice with intratracheal LPS (60 μg/head) treatment (control: n = 5, AEC-Reg1-cKO: n = 3). e RT-qPCR analysis for the indicated genes in the lung of WT C57BL/6J mice at 48 h after intratracheal administration of PBS or heat-killed P. aeruginosa (PA) (1.0 × 107 cfu/head) (n = 6 for each group). Data are shown as mean ± s.d. Two-tailed Student’s t-test was used for statistical analyses (b, d, e)
Among them, Pigr (polymeric immunoglobulin receptor; pIgR) mediates transcytosis, a function of AEC to capture circulating IgA and transport it to the apical side of the cell for the efficient secretion of IgA to the airway lumen.27 pIgR is expressed on the basal side of AECs, captures circulating IgAs followed by the active transport of the IgA-pIgR complex into the apical area,27 thus contributes to the airway defense against microorganisms.30 Consistent with increased pIgR expression (Fig. 5a), immunohistochemistry examination revealed that AECs in Reg1–/– lung are intensely stained with IgA. These results suggest that increased pIgR expression is associated with enhanced IgA levels in the airway mucosa under Regnase-1 deficiency in AECs (Fig. 5c).
Moreover, Regnase-1 is found to suppress the expression of Ltf, Muc5b, and Sftpd, which functions as endogenous antibiotics.31,32,33 The upregulation of these genes was also observed in AEC-Reg1-cKO lung compared with controls after intratracheal LPS administration (Fig. 5d). Interestingly, the expression of these mRNAs was induced in the lung in response to heat-killed P. aeruginosa (Fig. 5e), concomitant with the disappearance of Regnase-1 by the treatment. These data indicate that the loss of Regnase-1 directly enhances the AEC barrier functions and transcytosis mimicking P. aeruginosa-induced responses, and Regnase-1 contributes to the enhanced bacterial clearance in Reg1-AEC-cKO mice.
Regnase-1 in AECs governs the neutrophil accumulation
Next, we evaluated the contribution of Regnase-1 in AECs for the regulation of neutrophils in the lung. Flow cytometry analysis revealed that neutrophils were highly accumulated in the lungs of Reg1–/– mice than in WT controls (Fig. 6a). Histologically, massive accumulation of immune cells was observed in the lungs of Reg1–/– mice (Fig. 6b). Together with the accumulation of neutrophils in the lung of AEC-Reg1-cKO mice (Fig. 2a) these results demonstrate that AECs regulate neutrophil accumulation in the lung via Regnase-1.
Regnase-1 in AECs governs the neutrophil accumulation. a Flow cytometry analysis for neutrophils in the lung from WT or Reg1–/– mice. Images are representative of three samples for each group. b Representative images of hematoxylin and eosin (H&E) staining of lung samples from WT (n = 4) and Reg1–/– (n = 6) mice. c, d RT-qPCR analysis of the indicated genes in the lung from control or AEC-Reg1-cKO mice without treatment (e, n = 6 for each group) or intratracheal LPS (60 μg/head) treatment (f, control: n = 5, AEC-Reg1-cKO: n = 3). e Representative flow cytometry analysis showing neutrophil populations or total neutrophil numbers in the lung (left) or BALF (right) of intratracheal LPS (60 μg/head)-treated AEC-Reg1-cKO or control mice for 48 h. Image is a representative or sum of n = 3 for each group. f Ratio of CXCR2-blocked and non-blocked neutrophils in the lung and BALF after the adaptive transfer of fluorescence-labeled CD45.1 WT neutrophils to Regnase-1–/– mice (n = 6). Data are shown as mean ± s.d. Two-tailed Student’s t-test was used for statistical analyses (c–f)
We have previously reported that Cxcl1, a pivotal attractant of neutrophils into the airway, is a target of Regnase-1.20 Concordantly, we found that Cxcl1 mRNA levels are significantly elevated in the lung of AEC-Reg1-cKO mice compared with control (Fig. 6c), suggesting that CXCL1 could be one factor that contributes to the neutrophil accumulation observed under Regnase-1 deficiency. On the other hand, the enhancement of Cxcl5 expression was marginal under steady state (Fig. 6c). In response to intratracheal LPS instillation, the expression of chemokine genes such as Cxcl1, and Cxcl5 were further upregulated in AEC-Reg1-cKO mice compared with control (Fig. 6d). Consistent with the upregulation of Cxcl1 and Cxcl5, neutrophil populations and numbers in BALF were significantly elevated (Fig. 6e). To investigate if the accumulation of neutrophils into the lung and BALF is mediated by CXCL1/CXCL5 and their cognitive receptor CXCR2, we isolated neutrophils from the bone marrow (BM) of CD45.1 congenic mice and treated them with anti-CXCR2 neutralizing Ab or control IgG; then the cells were competitively transferred into Reg1–/– mice. The accumulation of neutrophils into the lung and BALF from the blood was significantly attenuated by the blockade of CXCR2 (Fig. 6f), indicating that CXCR2 signaling contributes to the recruitment of neutrophils to the lung and BALF under Regnase-1 deficiency.
Epithelial and immune cell Regnase-1 coordinates optimal immune response
Besides the enhancement of transcytosis, Regnase-1 in AECs was found to regulate IgA+ plasma cells in the lung following P. aeruginosa infection (Fig. 3c–e). Interestingly, analysis of the transcriptome data identified Ccl28, a critical chemokine for IgA+ plasma cell infiltration, as the Regnase-1 regulated gene in AEC (Fig. 4b). We confirmed that Ccl28 mRNA levels were significantly upregulated in the lung of both Reg1–/– mice and AEC-Reg1-cKO mice compared with control (Fig. 7a), suggesting that Regnase-1 in AECs controls plasma cell recruitment via Ccl28. However, we found no increase in both total and IgA+ plasma cell numbers in the lung (Supplementary Fig. 7a, b) or in BALF IgA concentration (Supplementary Fig. 7c) in AEC-Reg1-cKO mice under steady-state conditions, suggesting that activation of lymphoid cells is required to upregulate IgA+ cells.
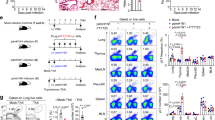
Epithelial and immune cell Regnase-1 coordinates optimal induction of acquired immunity. a RT-qPCR analysis of Ccl28 in the lung from WT and Reg1–/– mice or control and AEC-Reg1-cKO mice (n = 6). b, c Plasma cells (b) or IgA+ plasma cells (c) populations in the lung from T-Reg1-cKO BM transferred control (n = 4) and AEC-Reg1-cKO (n = 5) mice. d Rate of plasma cells among lymphocytes in the lung of T-Reg1-cKO BM transferred control and AEC-Reg1-cKO mice. e RT-qPCR analysis of Ccl20 in the lung from control and AEC-Reg1-cKO mice (n = 6). f, g Representative flow cytometry image (f) and average ratio (g) of Th17 population among CD4+ T cells in the lung of T-Reg1-cKO BM transferred control (n = 4) or AEC-Reg1-cKO mice (n = 5). h Venn diagram for the numbers of commonly increased mRNAs in Regnase-1-deficient CD4+ T cells and AECs. i GOs enriched in Regnase-1-deficient CD4+ T cells and AECs. GOs commonly enriched (blue bars), specifically enriched in Regnase-1-deficient T cells (green bars) or AECs (red bars) are shown. Data are shown as mean ± s.d. Two-tailed Student’s t-test was used for statistical analyses (a, d, f, g)
We previously showed that Regnase-1 expressed in T cells is critical for controlling the plasma cell number in lymphoid organs as well as serum Ab levels.19 Thus, we hypothesized that Regnase-1 in immune and non-immune cells coordinately governs airway IgA secretion. Indeed, analysis of CD4-Cre+ Regnase-1flox/flox mice (T-Reg1-cKO mice) and their littermate control mice at the age of 5–6 weeks revealed that there was a massive increase of plasma cells and IgA+ plasma cells in the mLN (Supplementary Fig. 8a, b). In contrast, the increase of plasma cells or IgA+ plasma cells in the lung was marginal (Supplementary Fig. 8a, b), which suggests that lack of Regnase-1 in T cells alone was not sufficient to populate plasma cells in the peripheral tissue (Supplementary Fig. 8b). In addition, B cell-specific Regnase-1-deficient mice generated by crossing CD19-Cre and Regnase-1flox/flox mice (B-Reg1-cKO) failed to increase plasma cells both in the lung and mLN (Supplementary Fig. 8c–e), indicating that Regnase-1 in B cells alone cannot explain the phenotype. We further tested the contribution of hematopoietic cells by generating BM chimeric mice lacking Regnase-1 in hematopoietic cells. We lethally irradiated CD45.1 WT mice, and transferred BM cells from WT (WT-BMT) or Reg1–/– (KO-BMT) mice (Supplementary Fig. 9a, b). A clear increase of plasma cells was observed in KO-BMT mice in mLN, but not in the lung (Supplementary Fig. 9c, d). These results demonstrate that the lack of Regnase-1 in lymphoid cells alone was not sufficient to populate plasma cells to the lung.
In order to address the possibility that Regnase-1 regulates the coordination of AECs and T cells for optimal induction of plasma cells in the lung, we transferred BM cells from T-Reg1-cKO mice into either AEC-Reg1-cKO mice or control mice. Lack of Regnase-1 both in AECs and T cells significantly increased the plasma cell population and IgA+ plasma cells in the lung (Fig. 7b–d), suggesting that T cell-mediated plasma cell differentiation and AEC-mediated plasma cell attraction coordinately act to optimally induce airway IgA secretion via Regnase-1 (Supplementary Fig. 10).
Regnase-1 in T cells has been shown to suppress Th17, Th1, and Th2 differentiation.19,23 Consistently, populations of Regnase-1-deficient T cells expressing transcription factors Rorγt, T-bet, and Gata3, but not Foxp3, were increased in the lung (Supplementary Fig. 11a, b), together with the enhanced property to produce cytokines, including IL-17, IFN-γ, and IL-4 (Supplementary Fig. 11c, d). Since IL-17 is known to activate AECs,34,35,36 we hypothesized that Regnase-1 in T cells and AECs collaborate to control respiratory Th17. Indeed, T-Reg1-cKO mice showed enhanced expression of the AEC-specific genes, suggesting that Regnase-1 in T cells controls immune effector functions to control AECs (Supplementary Fig. 11e). On the other hand, lack of Regnase-1 in AEC alone failed to increase Th17 cells in steady state (Supplementary Fig. 11f), but the Th17 population was significantly increased after heat-killed P. aeruginosa treatment (Supplementary Fig. 11g), consistent with the enhanced expression of Ccl20, a chemokine that attracts Th17 to the mucosal area (Figs. 4b, 7e, and Supplementary Fig. 5). Whereas γδT cells are also known to produce IL-17, the ratio of Th17 cells in the IL-17-producing cells were much more than γδT cells in AEC-Reg1-KO mice following heat-killed P. aeruginosa treatment (Supplementary Fig. 11h). These data indicate that the loss of Regnase-1 in AECs facilitates the accumulation of Th17 into the lung to produce IL-17. Interestingly, lack of Regnase-1 both in AECs and T cells led to a significant increase of Th17 cells in the lung (Fig. 7f, g). Collectively, these results indicate the bidirectional regulatory mechanism between AECs and T cells in accumulation of IgA-producing cells and Th17 cells, which further govern effector immune reactions.
To further dissect the mechanism how Regnase-1 controls immune-related functions in distinct cell types, we analyzed the transcriptome data in Regnase-1-deficient AECs (Fig. 4a) and CD4+ T cells.19 By comparing genes with significant upregulation under Regnase-1 deficiency (>2-fold upregulation with p < 0.05) in CD4+ T cells (n = 1459) or AECs (n = 480), we found that only few genes (n = 59) overlapped (Fig. 7h), indicating that Regnase-1 targets almost exclusive sets of genes in T cells and AECs. However, interestingly, gene ontology (GO) analysis revealed that Regnase-1-regulated genes in AECs and T cells share a set of common GO terms related with immune reactions in addition to a set of cell-specific GO terms (Fig. 7i). Collectively, these results suggest that Regnase-1 controls distinct sets of genes depending on the cell types, but the genes working for a common function, and participate in a multi-cellular immune network.
Regnase-1 directly interacts and degrades AEC-specific target mRNAs
Given that Regnase-1-controlled genes are distinct depending on cell types, we then examined if genes upregulated in Regnase-1-deficient AECs are directly controlled by Regnase-1 by RNA-immunoprecipitation (RIP)-qPCR analysis. To immunoprecipitate Regnase-1 in vivo, we generated mice expressing the FLAG-tag in the C-terminal end of Regnase-1 (Reg1-FLAG-KI mouse) by using the CRISPR/Cas9 system (Supplementary Fig. 12a). The expression of Regnase-1 was not affected by the insertion of FLAG-tag in the C-terminal portion (Supplementary Fig. 12b), and Regnase-1 was successfully immunoprecipitated with anti-FLAG Ab (Supplementary Fig. 12b, c). By using the lysate from the lung of Reg1-FLAG-KI and WT mouse, we found that FLAG-Regnase-1 co-precipitated mRNAs regulated by Regnase-1 in AECs including Pigr, Ccl28, Cxcl5, Ltf, Muc5b, but not Nfkbia (Fig. 8a), confirming the direct interaction between Regnase-1 and these mRNAs. Then we examined if Regnase-1 directly degrade interacting mRNAs. By using the Tet-off system in HEK293 cells, we found that Pigr or Ccl28 mRNA are destabilized by the overexpression of WT, but not nuclease-dead mutant (D141N), Regnase-1 (Fig. 8b). When we expressed luciferase reporter constructs harboring 3′UTR of genes including Ltf, Muc5b, Cxcl5, Ccl20, Sftpd, Pigr, and Ccl28 together with WT or D141N Regnase-1, the luciferase activity was significantly suppressed in response to the expression of WT, but not D141N, suggesting that these genes are regulated by Regnase-1 through 3′UTRs (Fig. 8c). Collectively, these data showed that Regnase-1 directly destabilizes AEC-specific target mRNA through its RNase activity.
Regnase-1 directly destabilizes AEC-specific target mRNAs. a RNA-immunoprecipitation (RIP)-qPCR analysis of indicated genes in the lung of WT or Reg1-FLAG-Knock-in (Reg1-FLAG-KI) mouse. Relative enrichment of mRNA amount compared with input sample is shown. Each dot indicates one sample. b Tet-off assay to analyze the altered stability of Ccl28 or Pigr mRNA in the presence of Regnase-1. HEK293 tet-off cells were co-transfected with either of WT or D141N mutant Regnase-1-expressing plasmid or control (empty) plasmid together with pTREtight-expressing CDS plus 3′UTR sequences of the genes indicated. The amount of remaining RNA was evaluated by RT-qPCR, and the amount at 2 h was compared. Each value indicates the average of three independent experiments. c Luciferase reporter assays for the evaluation of 3′UTR-dependent suppression of gene expression by Regnase-1. HEK293 cells were co-transfected with WT or D141N mutant Regnase-1-expressing plasmid or control (empty) plasmid together with pGL3-promotor plasmid with the 3′UTR sequence of the indicated genes, and luciferase activity was evaluated 48 h after the transfection. Data are representative of two independent experiments with n = 3. Data are shown as mean ± s.d. Two-tailed Student’s t-test was used for statistical analyses (b, c)
Discussion
In the present study, we showed that AECs regulate surface barrier functions and immune responses against P. aeruginosa infection in the lung by Regnase-1-mediated post-transcriptional control. Regnase-1 in the lung undergoes degradation in response to the exposure to bacteria and the disappearance was sustained following treatment with P. aeruginosa. AEC-specific deletion of Regnase-1 resulted in the highly improved resistance against virulent P. aeruginosa infection of the airway, particularly in response to re-infection. The biological effects of Regnase-1 degradation by infection include the following: (1) upregulation of endogenous anti-microbial components; (2) accumulation of neutrophils; and (3) enhancement of IgA secretion. These effects are optimized through coordination between AECs and other professional immune cells including T cells. Regnase-1 governs this coordination by regulating distinct gene sets in each cell type.
Recognition of pathogens by TLRs in sensor cells induces gene expression to promote innate immune responses to clear infections by transcriptional and post-transcriptional regulations. We found that in vivo pulmonary treatment with P. aeruginosa induced sustained degradation of Regnase-1, whereas NF-κB activation was induced only transiently. These results suggest that Regnase-1, rather than the NF-κB-mediated transcriptional program, plays a critical role in sustaining innate responses in the lung in response to bacterial infection. We have previously shown that Regnase-1 degradation is induced by IKK-mediated phosphorylation downstream of the TLR signaling.21 Since IKK is essential for NF-κB activation, it was postulated that NF-κB and Regnase-1 are co-regulated in innate immune cells in response to TLR. Interestingly, the BCL10-MALT1 complex was also found to regulate the protein level of Regnase-1 in the lung, especially at the later phase of inflammation. We have previously shown that the BCL10-MALT1 is important for the degradation of Regnase-1 in T cells in the TCR signaling. However, it has been reported that the BCL10-MALT1 complex is involved in the signaling downstream of not only TCR, but also C-type lectin receptors, Fc receptors, and G-protein-coupled receptors. Thus, the degradation of Regnase-1 is possibly mediated by the signaling of multiple sensors recognizing P. aeruginosa to activate both MyD88 and BCL10-MALT1. Further studies are required to uncover combinations of immune sensors controlling Regnase-1 expression operating in the lung following bacterial infection.
AECs not only act as a physical barrier in the airway but also secrete various endogenous antimicrobials and produce cytokines and chemokines to activate phagocytes, thus contributing to the first-line defense against pathogens. Regnase-1 expressed in AECs specifically regulates genes, such as Ltf, Muc5b, and Sftpd, whose expression is not controlled in immune cells. These endogenous antimicrobials function in distinct mechanisms. For instance, Lactoferrin binds iron competitively with bacteria, thus inhibiting bacterial growth.31 MUC5B traps bacteria, and exclude them in coordination with ciliary movement.33 Surfactant protein D not only functions as a direct antimicrobial, but opsonizes bacteria for phagocytes.32 pIgR is also expressed specifically in mucosal epithelial cells. Given that these mRNAs are directly regulated by Regnase-1 potentially through stem-loop structures in their 3′UTRs, it is intriguing that these antimicrobials developed to harbor Regnase-1-regulatory elements in the mRNAs. In addition to Regnase-1, immune cell activation is known to be controlled by RNA-binding proteins such as tristetraprolin (TTP) and Roquin.13,15,16,22,37 TTP destabilizes the mRNA through the deadenylation by recognizing AU-rich element in the 3′UTR of its target mRNA such as TNF.38,39 Although TTP is reported to destabilize IL-8 and TNF mRNAs in AECs similar to immune cells,40,41 AU-rich elements were not found in mRNAs encoding antimicrobials and pIgR. Although Roquin 1/2 recognizes stem-loop structures in mRNA 3′UTRs that overlap with those of Regnase-1,20 it was reported that Sanroque mutant mice harboring a dysfunctional mutation in Roquin-1 did not show severe lung inflammation like that observed under Regnase-1 deficiency.42 Regnase-1 is in fact the first example of a molecular switch in AECs that controls lung barrier functions as well as immune responses dynamically regulated in the course of pulmonary infection. Regnase-1 can prevent overactivation of the immune system while minimizing disturbance of rapid induction of target genes.
Regnase-1 expressed in AECs is critical for controlling the resistance against P. aeruginosa re-infection in vivo. Interestingly, local production and secretion of IgA to the airway mucosa as well as accumulation of Th17 in the lung were enhanced by the lack of Regnase-1 in AECs. These results indicate that AECs contribute not only to earliest stage cell-intrinsic innate immunity but also to the control of lymphocyte-mediated second tier immunity via Regnase-1. In contrast, AECs in Regnase-1 does not suppress systemic acquired immune responses. Therefore, AEC-mediated activation of lymphocytes via Regnase-1 might be specialized to control local immune responses without affecting systemic activation. Regnase-1-mediated control of chemokines such as CCL20 and CCL28 possibly regulates lymphocytes accumulation in the lung and local immune responses, though further studies are required to identify precise mechanisms how AECs control acquired immune reactions in the lung.
The present study also showed that Regnase-1 governs the coordination between AECs and T cell-mediated induction of acquired immunity. We have previously shown that TCR-mediated T cell activation in lymph nodes induced the degradation of Regnase-1, contributing to plasma cell differentiation.19 On the other hand, degradation of Regnase-1 in AEC contributes to the optimal enhancement of airway IgA secretion and acquired resistance against pathogens such as P. aeruginosa. Interestingly, we found that loss of Regnase-1 both in AECs and T cells resulted in the increase of Th17 cells in the lung. IL-17 directly activates AECs,35,43 and this activation is required for protection against gram-negative bacterial infection.36 Notably, AEC-specific Regnase-1 target genes such as Pigr, Lcn2, Cxcl1, Cxcl5, and Ccl20 are also known as IL-17-inducible genes in AECs.34,36 It was reported that Regnase-1 amplifies IL-17-mediated signaling in skin tissues.44 Indeed, we found that the genes upregulated in AECs or T cells in the absence of Regnase-1 largely share gene ontologies related to the regulation of immunity by comparing transcriptome analysis. On the other hand, we found that only few genes were commonly regulated by Regnase-1 both in AECs and T cells. Although it is not clear how Regnase-1 controls distinct sets of mRNAs depending on cell types, it is possible that cell-type-specific expression of gene expression is regulated by a combination of transcriptional-mediated and mRNA-mediated control. Indeed, NF-κB-binding sites were identified in PIGR and MUC5B genes and are thought to be involved in the regulation of their expression.27,45 Although the NF-κB signaling pathways are activated in various cell types in response to TLR stimulation, interaction with tissue-specific transcription factors and/or epigenetic regulation might explain the tissue-specific regulation of mRNA stability by Regnase-1.
An open question is how T cells and AECs cooperate to induce AEC-specific Regnase-1 target genes. Although IL-17 is an obvious candidate, our initial attempt to neutralize IL-17 signaling by treating heat-killed P. aeruginosa-stimulated AEC-Reg1-cKO mice with anti-IL17RC Ab failed to decrease the expression levels of Pigr, Cxcl1, Ccl28, and Cxcl5; however, the Muc5b mRNA levels appeared to be suppressed (data not shown). These results suggest that IL-17R signaling modestly contributes to the enhancement of Regnase-1-target mRNA expression under Regnase-1 deficiency in AECs. Thus, the direct regulation of mRNAs such as Pigr by Regnase-1 in an AEC-intrinsic fashion might be more important for the control of such mRNA expression. In contrast, we also found that T-Reg1-cKO mice expressed higher amounts of Pigr mRNA (Supplementary Fig. 11e), indicating that T cells lacking Regnase-1 harbor the potential to upregulate Regnase-1 target genes in AECs. Recently, it was reported that IL-17A stimulation induced the degradation of Regnase-1 in keratinocytes.46 Thus, it is tempting to speculate that Regnase-1 is also degraded by IL-17 stimulation in AECs, which might explain the effect of IL-17 in controlling gene expression in AECs. Alternatively, Th subsets other than Th17, which are increased in the absence of Regnase-1 in T cells, might contribute to the induction of Regnase-1-target mRNAs in AECs. Future analysis will be required to uncover the role of IL-17 and other Th cytokines in the regulation of Regnase-1-mediated expression in AECs.
The present study offers several potential benefits for the inhibition of AEC Regnase-1 in a clinical setting. First, it may offer an alternative therapeutic option for pneumonia. For example, P. aeruginosa is often resistant to antibiotic drugs, and that makes the treatment of pneumonia difficult.1,2 In this situation, Regnase-1 inhibition may enhance endogenous property for elimination of pathogens. Second, suppression of Regnase-1 possibly functions as an adjuvant for vaccination. Vaccines currently available for airway infections such as influenza virus have some limitation in their efficacy to prevent infection. For example, these vaccines induce IgG, but IgG is not effectively transported to the airway space. In addition, because of the high antigen specificity of the IgG induced by these vaccines, they might not offer protection against pathogens with minor mutations. In contrast, IgA is more effectively secreted into the airway space by the help of pIgR, and it was reported that airway-secreted IgA can be protective against even diverse antigens,47 indicating that an IgA-inducible vaccine may have a greater potential to prevent infection. Unfortunately, IgA-inducible vaccines have not reached clinical efficiency yet,48 so inhibition of Regnase-1 in AECs coupled with antigen may enhance protective IgA secretion.
In conclusion, we have shown that Regnase-1 in AECs plays a significant role in the protection against pathogens. This process is initiated by TLR-mediated degradation of Regnase-1, and coordination of antibiotic secretion, neutrophil induction, and Th17 recruitment enables rapid elimination of pathogens. In addition, the enhancement of late-phase pathogen-specific IgA secretion may contribute to protection against subsequent infections. These results offer a potential clinical benefit of airway epithelial Regnase-1 inhibition as a novel strategy for the treatment and the prevention of pulmonary infection.
Methods
Mice
C57BL/6J mice were purchased from CLEA Japan, Inc. (Tokyo, Japan). Reg1–/–, Regnase-1flox/flox, MyD88–/–, CD4-Cre transgenic, CD19-Cre transgenic, Sanroque mice have been previously described.18,19,49,50 Nkx2.1-Cre transgenic mice and ROSA26mT/mG mice26 were purchased from Jackson Laboratory (Bar Harbor, ME, USA). All mice were raised under specific pathogen-free condition except for the infection experiments. As for AEC-Reg1-cKO mice, Nkx2.1-Cre+ Regnase-1flox/+ mice or Regnase-1flox/– mice were utilized for control. As for T-Reg1-cKO mice, CD4-Cre+ Regnase-1flox/+ mice were utilized for control. All protocols of animal experiments were approved by the Kyoto University Animal Experimentation Committee.
Cells and transfection
HEK293 cells were maintained in DMEM (Nacalai Tesque) supplemented with 10% FBS, 50 μM β-mercaptoethanol (Invitrogen). Calu-3 cells were maintained in MEM (Nacalai Tesque) supplemented with 10% FBS, 1 mM sodium pyruvate, 50 μM β-mercaptoethanol (Invitrogen). Tet-off HEK293 cells were purchased from Clontech and maintained in α-MEM (Nacalai Tesque) supplemented with 10% FBS (Clontech), 50 μM β-mercaptoethanol (Invitrogen), and 100 μg/ml G-418 (Nacalai Tesque). Plasmids were transfected into cells with the use of Lipofectamine 2000, Lipofectamine 3000 (Invitrogen), or the Neon transfection system (Invitrofen) according to the manufacturer’s instruction.
Preparation of single-cell suspension from the lung
Murine lung samples were minced with scissors and digested in DNase I (Roche applied science) plus Collagenase D (Roche applied science) containing RPMI medium for 50 min at 37 °C. Then 500 mM EDTA was added as to the final concentration of 10 mM, and incubated for 10 min at 37 °C. Following to the digestion, the digested lung tissue was filtered with 40 μm cell strainer, and the cells were collected. For the isolation of lung epithelial cells, filtered lung tissue was further digested by GentleMACS™ Dissociator (Miltenyi Biotec), filtered again, and cells were collected.
FACS analysis
For FACS analyses, cells were incubated with Zombie NIR solution (BioLegend) on ice for 10 min, anti-mouse CD16/CD32 (BioLegend) on ice for 15 min, and stained with Ab cocktail as indicated on ice for 20 min. For the analysis of cytokines in T cells, the cells were re-stimulated for 4 h with 50 ng/ml phorbol-12-myristate-13-acetate (PMA) and 1 μM ionomycin in the presence of GolgiStopTM (BD Biosciences). For intracellular staining, cells were fixed with Fixation/Permeabilization solution (eBioscience) or Lyse/Fix buffer (BD Biosciences) stained with Ab cocktail suspended with either of permeabilization buffer (eBioscience) or Perm/Wash buffer I (BD Biosciences), according to the manufacturer’s indication. The cells were washed, and subjected to the analyses. The following Abs are used: FITC-conjugated anti-mouse IgA (C10-3, BD Pharmingen), FITC-conjugated anti-mouse Gr-1(RB6-8C5, BioLegend), PE-conjugated anti-mouse CD138 (281-2, BioLegend), PE-conjugated anti-mouse IL-17A (TC11-18H10, BD Biosciences), PE-conjugated anti-mouse GATA3 (TWAJ, Thermo Fisher), PE-conjugated anti-mouse IL-4, APC-conjugated anti-mouse CD19 (1D3, BD Pharmingen), APC-conjugated anti-mouse CD11b (M1/70, BioLegend), APC-conjugated anti-mouse CCR6 (29-2L17, BioLegend), APC-conjugated anti-mouse CD45 (30-F11, Tonbo Biosciences), APC-conjugated anti-mouse T-bet (4B10, BioLegend), APC-conjugated anti-mouse Rorγt (B2D, eBioscience), PerCP-conjugated anti-mouse CD4 (GK1.5, BioLegend), PE-Cy7-conjugated anti-mouse TCRβ (H57-597, BioLegend), PE-Cy7-conjugated CD45.1 (A20, BioLegend) and PE-Cy7-conjugated CD45.2 (104, BioLegend). Dead cells were recognized by Zombie-NIR or Zombie-Aqua (BioLegend). FACS data were obtained by FACSVerse (BD Biosciences) or LSR Fortessa X-20 (BD Biosciences), and analyzed by using FlowJo software (FlowJo, LLC).
Intratracheal administration of lipopolysaccharide or P. aeruginosa
P. aeruginosa strain PA103 was cultured on tryptic soy agar plate (Sigma). Colonies were picked up and cultured with tryptic soy broth. Cfu for P. aeruginosa was estimated by turbidity OD620. For the injection of live PA103, the cultured medium was centrifuged at 2000 × g for 5 min, washed with PBS three times, and suspended with PBS up to the indicated concentration. Heat-killed P. aeruginosa was prepared by heating concentration-determined PA103 suspension at 95 °C for 30 min. For the experiments with WT mice, 8-weeks-old female C57BL/6J mice were utilized. For the experiments with AEC-Reg1-cKO and control mice, sex and age (between 7 and 20 wk old) were matched between both groups. Mice were anesthetized with intraperitoneal injection of pentobarbital with inhalation of volatile anesthesia, and trachea was exposed by small incision. Trachea was punctured with 27G needle, and the indicated dose of LPS or P. aeruginosa suspended in 20 μl PBS was administered. For the neutralization of IL-17RC, 100 μg of anti-IL-17RC Ab (Novus Biologicals, AF2270) was suspended by PBS and administered simultaneously with heat-killed P. aeruginosa.
Immunoblot
Cell lysate was prepared with RIPA buffer. Immunoblot was performed as described previously.20 The following Abs were used: anti-mouse pIgR (AF2800, R&D), anti-mouse MUC5B, anti-mouse Surfactant protein D, and anti-mouse/human β-actin (sc-1615; Santa Cruz). Rabbit anti-mouse/human Regnase-1 Ab has been described previously.21 Luminescence was detected with a luminescent image analyzer (ImageQuant LAS 4000; GE Healthcare), and then analyzed by using ImageJ software (NIH, Bethesda, Maryland).
RNA sequencing and transcriptome analysis
Cultured MTEC from WT or Reg1–/– mice were lysed with TRIzol (Invitrogen), and total RNA was extracted as described above (duplicated samples). It was confirmed that RNA integrity number for each sample was more than 9.0, and then the samples were sequenced on a HiSeq 2000 system (Illumina) according to the manufacturer’s instruction. Identified reads were mapped on the murine genome (GRCm38) using HISAT2 (version 2.1.0),51 and the number of reads mapped to each gene determined using HTSeq52 (using option --stranded=no) and the GRCm38 transcriptome gene annotation (.gtf file, Ensembl release 84). The P-value for the difference of expression level and log2 fold change in normalized read count of each gene in Reg1–/– MTEC compared with WT were evaluated by using DESeq2.53 GO analysis was performed by using GOstats,54 and ranked according to the P-value.
Statistical analysis
Data are presented as mean ± standard deviation. Student’s t-test or Mann–Whitney’s U test were used for the analysis of continuous data. Wilcoxon test was used for the analysis of survival. A P-value of <0.05 was regarded as significant.
Accessioncodes
DNA Data Bank of Japan (DDBJ): DRA006334.
References
Tumbarello, M. et al. Clinical outcomes of Pseudomonas aeruginosa pneumonia in intensive care unit patients. Intens. Care Med. 39, 682–692 (2013).
Park, Y. S. et al. Acquisition of extensive drug-resistant Pseudomonas aeruginosa among hospitalized patients: risk factors and resistance mechanisms to carbapenems. J. Hosp. Infect. 79, 54–58 (2011).
Iwasaki, A., Foxman, E. F. & Molony, R. D. Early local immune defences in the respiratory tract. Nat. Rev. Immunol. 17, 7–20 (2017).
Iwasaki, A. & Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16, 343–353 (2015).
Hammad, H. & Lambrecht, B. N. Barrier epithelial cells and the control of type 2 immunity. Immunity 43, 29–40 (2015).
Kato, A. & Schleimer, R. P. Beyond inflammation: airway epithelial cells are at the interface of innate and adaptive immunity. Curr. Opin. Immunol. 19, 711–720 (2007).
Whitsett, J. A. & Alenghat, T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 16, 27–35 (2015).
Bartlett, J. A., Fischer, A. J. & McCray, P. B. Jr. Innate immune functions of the airway epithelium. Contrib. Microbiol. 15, 147–163 (2008).
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
Sha, Q., Truong-Tran, A. Q., Plitt, J. R., Beck, L. A. & Schleimer, R. P. Activation of airway epithelial cells by toll-like receptor agonists. Am. J. Respir. Cell Mol. Biol. 31, 358–364 (2004).
Blackwell, T. S., Blackwell, T. R., Holden, E. P., Christman, B. W. & Christman, J. W. In vivo antioxidant treatment suppresses nuclear factor-kappa B activation and neutrophilic lung inflammation. J. Immunol. 157, 1630–1637 (1996).
D’Alessio, F. R. et al. CD4+CD25+Foxp3+ Tregs resolve experimental lung injury in mice and are present in humans with acute lung injury. J. Clin. Invest. 119, 2898–2913 (2009).
Anderson, P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat. Rev. Immunol. 10, 24–35 (2010).
Mino, T. & Takeuchi, O. Post-transcriptional regulation of cytokine mRNA controls the initiation and resolution of inflammation. Biotechnol. Genet. Eng. Rev. 29, 49–60 (2013).
Kafasla, P., Skliris, A. & Kontoyiannis, D. L. Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat. Immunol. 15, 492–502 (2014).
Athanasopoulos, V., Ramiscal, R. R. & Vinuesa, C. G. ROQUIN signalling pathways in innate and adaptive immunity. Eur. J. Immunol. 46, 1082–1090 (2016).
Fu, M. & Blackshear, P. J. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat. Rev. Immunol. 17, 130–143 (2017).
Matsushita, K. et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458, 1185–1190 (2009).
Uehata, T. et al. Malt1-induced cleavage of Regnase-1 in CD4(+) helper T cells regulates immune activation. Cell 153, 1036–1049 (2013).
Mino, T. et al. Regnase-1 and roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell 161, 1058–1073 (2015).
Iwasaki, H. et al. The IkappaB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of Regnase-1. Nat. Immunol. 12, 1167–1175 (2011).
Jeltsch, K. M. et al. Cleavage of roquin and Regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat. Immunol. 15, 1079–1089 (2014).
Garg, A. V. et al. MCPIP1 endoribonuclease activity negatively regulates interleukin-17-mediated signaling and inflammation. Immunity 43, 475–487 (2015).
DiMango, E., Ratner, A. J., Bryan, R., Tabibi, S. & Prince, A. Activation of NF-kappaB by adherent Pseudomonas aeruginosa in normal and cystic fibrosis respiratory epithelial cells. J. Clin. Invest. 101, 2598–2605 (1998).
Tiozzo, C. et al. Deletion of Pten expands lung epithelial progenitor pools and confers resistance to airway injury. Am. J. Respir. Crit. Care Med. 180, 701–712 (2009).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Kaetzel, C. S. The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunol. Rev. 206, 83–99 (2005).
Morteau, O. et al. An indispensable role for the chemokine receptor CCR10 in IgA antibody-secreting cell accumulation. J. Immunol. 181, 6309–6315 (2008).
Yamazaki, T. et al. CCR6 regulates the migration of inflammatory and regulatory T cells. J. Immunol. 181, 8391–8401 (2008).
Richmond, B. W. et al. Airway bacteria drive a progressive COPD-like phenotype in mice with polymeric immunoglobulin receptor deficiency. Nat. Commun. 7, 11240 (2016).
Ganz, T. Antimicrobial polypeptides. J. Leukoc. Biol. 75, 34–38 (2004).
Pastva, A. M., Wright, J. R. & Williams, K. L. Immunomodulatory roles of surfactant proteins A and D: implications in lung disease. Proc. Am. Thorac. Soc. 4, 252–257 (2007).
Roy, M. G. et al. Muc5b is required for airway defence. Nature 505, 412–416 (2014).
Aujla, S. J. et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 14, 275–281 (2008).
Jaffar, Z., Ferrini, M. E., Herritt, L. A. & Roberts, K. Cutting edge: lung mucosal Th17-mediated responses induce polymeric Ig receptor expression by the airway epithelium and elevate secretory IgA levels. J. Immunol. 182, 4507–4511 (2009).
Chen, K. et al. IL-17 receptor signaling in the lung epithelium is required for mucosal chemokine gradients and pulmonary host defense against K. pneumoniae. Cell Host Microbe 20, 596–605 (2016).
Pratama, A. et al. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity 38, 669–680 (2013).
Taylor, G. A. et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4, 445–454 (1996).
Marchese, F. P. et al. MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J. Biol. Chem. 285, 27590–27600 (2010).
Smoak, K. & Cidlowski, J. A. Glucocorticoids regulate tristetraprolin synthesis and posttranscriptionally regulate tumor necrosis factor alpha inflammatory signaling. Mol. Cell. Biol. 26, 9126–9135 (2006).
Balakathiresan, N. S. et al. Tristetraprolin regulates IL-8 mRNA stability in cystic fibrosis lung epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 296, L1012–L1018 (2009).
Nagalingam, G., Vinuesa, C. G., Britton, W. J. & Saunders, B. M. Modulation of roquin function in myeloid cells reduces Mycobacterium tuberculosis-induced inflammation. J. Immunol. 199, 1796–1804 (2017).
Iwakura, Y., Nakae, S., Saijo, S. & Ishigame, H. The roles of IL-17A in inflammatory immune responses and host defense against pathogens. Immunol. Rev. 226, 57–79 (2008).
Monin, L. et al. MCPIP1/Regnase-1 restricts IL-17A- and IL-17C-dependent skin inflammation. J. Immunol. 198, 767–775 (2017).
Fujisawa, T. et al. NF-kappaB mediates IL-1beta- and IL-17A-induced MUC5B expression in airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 45, 246–252 (2011).
Takaishi, M., Satoh, T., Akira, S., Sano, S. Regnase-1, an immuno-modulator, limits the IL-36/IL-36R auto-stimulatory loop in keratinocytes to suppress skin inflammation. J. Invest. Dermatol. (2018). https://doi.org/10.1016/j.jid.2017.12.033.
Tamura, S. et al. Cross-protection against influenza-a virus-infection by passively transferred respiratory-tract Iga antibodies to different hemagglutinin molecules. Eur. J. Immunol. 21, 1337–1344 (1991).
van Riet, E., Ainai, A., Suzuki, T. & Hasegawa, H. Mucosal IgA responses in influenza virus infections; thoughts for vaccine design. Vaccine 30, 5893–5900 (2012).
Rickert, R. C., Roes, J. & Rajewsky, K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25, 1317–1318 (1997).
Vinuesa, C. G. et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 435, 452–458 (2005).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Falcon, S. & Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 (2007).
Acknowledgements
We thank all members of our laboratory for discussions and M. Tsuji for secretarial assistance. We also thank Daron M. Standley (Research Institute for Microbial Diseases, Osaka University, Suita, Japan), Sarang Tartey (Department of Immunology, St. Jude Children’s Research Hospital, Memphis, TN, USA), Shigeo Muro, Hisako Matsumoto, Kiminobu Tanizawa, Shinpei Goto, Kohei Ikezoe, Koichi Hasegawa, Kiyoshi Uemasu (Department of Respiratory Medicine, Graduate School of Medicine, Kyoto University, Kyoto, Japan), and Michiaki Mishima (Saiseikai Noe Hospital, Osaka, Japan) for discussions and kind instructions for experimental procedures. This work was supported by the JSPS through Core-to-Core Program, Grant-in-Aid for Scientific Research (C) (to T.M.), Grant-in-Aid for JSPS Research Fellow (to Y.N.), for Scientific Research on Innovative Areas “Genome Science” (221S0002 and 16H06279), and grants from Takeda Science Foundation, Daiichi Sankyo Foundation of Life Science, Uehara Foundation (to O.T.), and the Shimizu Foundation for Immunology and Neuroscience (to T.M. and T.U.).
Author information
Authors and Affiliations
Contributions
All authors participated in the discussion and interpretation of the study. N.Y., T.M., T.U., and O.T. designed the experiments. N.Y. performed the majority of experiments and analysis of data. T.M., T.U., M.Y., X.C. helped with experiments. Y.S. performed RNA sequencing and the analysis of its data. A.V. performed detailed analysis of RNA sequence data. T.T. performed histological analysis. A.S. supported the detailed analysis of the cells in the lung. T.S. helped with infection experiments. N.Y. and O.T. wrote the manuscript. O.T. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Nakatsuka, Y., Vandenbon, A., Mino, T. et al. Pulmonary Regnase-1 orchestrates the interplay of epithelium and adaptive immune systems to protect against pneumonia. Mucosal Immunol 11, 1203–1218 (2018). https://doi.org/10.1038/s41385-018-0024-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-018-0024-5
This article is cited by
-
Regulation of inflammatory diseases via the control of mRNA decay
Inflammation and Regeneration (2024)
-
Innate immune sensing of pathogens and its post-transcriptional regulations by RNA-binding proteins
Archives of Pharmacal Research (2023)
-
The nucleolus is the site for inflammatory RNA decay during infection
Nature Communications (2022)
-
Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis
Nature (2020)