Abstract
Mucosal-associated invariant T (MAIT) cells are abundant innate-like T lymphocytes in mucosal tissues and recognize a variety of riboflavin-related metabolites produced by the microbial flora. Relevant issues are whether MAIT cells are heterogeneous in the colon, and whether the local environment influences microbial metabolism thereby shaping MAIT cell phenotypes and responses. We found discrete MAIT cell populations in human colon, characterized by the diverse expression of transcription factors, cytokines and surface markers, indicative of activated and precisely controlled lymphocyte populations. Similar phenotypes were rare among circulating MAIT cells and appeared when circulating MAIT cells were stimulated with the synthetic antigens 5-(2-oxoethylideneamino)-6-d-ribitylaminouracil, and 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil. Furthermore, bacteria grown in colon-resembling conditions with low oxygen tension and harvested at stationary growth phase, potently activated human MAIT cells. The increased activation correlated with accumulation of the above antigenic metabolites as indicated by mass spectrometry. Thus, the colon environment contributes to mucosal immunity by directly affecting bacterial metabolism, and indirectly controlling the stimulation and differentiation of MAIT cells.
Similar content being viewed by others
Introduction
Mucosal-associated invariant T (MAIT) cells are a unique subset of evolutionary-conserved innate-like T lymphocytes found in blood and mucosae. MAIT cells express a semi-invariant TCR1 with marked TCR oligoclonality.2 In humans, MAIT TCR is composed of three TCRα chains in which TRAV1-2 is rearranged to TRAJ33, TRAJ20, or TRAJ12 and paired with TCRβ chains mostly belonging to the TRBV6 and TRBV20 families.2,3,4 MAIT cells responding to diverse pathogens including Mycobacterium smegmatis, Salmonella typhimurium, and Candida albicans displayed pathogen-specific differences in their TCRβ usage.5 These findings support substantial heterogeneity across the MAIT cell repertoire and suggested the presence of adaptive-like responses within some MAIT cell populations.4,5,6
MAIT cells comprise 1–5% of T cells in the periphery and are present at ~2% and 10–30% of tissue resident T cells in mucosal sites and in the liver, respectively, in humans and mice.1,6,7,8 They develop in the thymus upon interaction with MHC class I-related protein (MR1) expressed on CD4+CD8+ thymocytes.9 Mature MAIT cells exit the thymus as naïve cells, seed bone marrow3 and require commensal flora and B cells for acquisition of effector/memory phenotype.1,10 The probable role of B cells is to present antigens to MAIT cells in defined niches, whereas that of commensal flora is to release antigenic molecules that stimulate MAIT TCR.11,12 Indeed, only infections with bacteria, which produce the stimulatory antigens, activate MAIT cells in vivo.13 In some instances MAIT cells are stimulated by an array of cytokines independently from TCR engagement.8,14,15
Metabolites generated in the riboflavin (vitamin B2) pathway stimulate MAIT cells.16,17,18,19 The key molecule is the intermediate metabolite 5-amino-6-(1-d-ribitylamino)uracil (5-A-RU), which gives rise to structurally related antigens. The first antigen is the enzymatically generated 6,7-dimethyl-8-(d-ribityl)lumazine16 (RL-6,7-diMe), the immediate precursor of riboflavin.20 This compound also gives rise to 7-hydroxy-6-methyl-8-d-ribityllumazine and to reduced 6-hydroxymethyl-8-d-ribityllumazine, which both stimulate MAIT cells.16
Other two antigens are pyrimidine-like molecules, namely 5-(2-oxoethylideneamino)-6-d-ribitylaminouracil (5-OE-RU) and 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil (5-OP-RU).18 These compounds result from non-enzymatic condensation of 5-A-RU with the dialdehyde glyoxal or the ketoaldehyde methylglyoxal18 (MG), respectively, and thus are adducts representing chemical side reactions of the pathway not required for riboflavin synthesis.
Both 5-OP-RU and 5-OE-RU, but not RL-6,7-diMe, make a Schiff base with Lys43 of MR1, generating highly stable complexes with MR1 and stabilizing it in a MAIT cell-stimulatory conformation.18 Increased expression of MR1 on the surface of antigen-presenting cells (APCs) pulsed with these antigens is a consequence of this stabilization.21,22 In contrast, RL-6,7-diMe binds weakly to MR1 and refolds inefficiently with recombinant MR1.17 MAIT cell activation is strongly influenced by the mode of ligand binding to MR1 and the chemical structure of the stimulatory metabolites. For example, 5-OP-RU has a very short half-life, and becomes stable only after binding MR1.22 Thus, the physiological relevance of 5-OP-RU is determined by immediate formation of a Schiff base with MR1. The variety of MAIT cell antigens raises questions about the nature of stimulatory metabolites produced under physiological conditions, and specificities of their effects on MAIT cell response.
The metabolism of commensal and opportunistic microbes in the gut constantly adapts to changing host conditions.23,24,25,26,27 Whether the mucosal physiological conditions affect the relative abundance of MAIT-stimulatory metabolites, thus influencing the type of MAIT cell activation, has not yet been investigated. Here we show that diverse metabolite antigens induce differential MAIT cell responses. In addition, we found that growth conditions affect the relative abundance of these MAIT cell-stimulatory compounds in bacteria, thereby influencing MAIT responses.
Results
MAIT cells freshly isolated from normal human colon exhibit activation signature
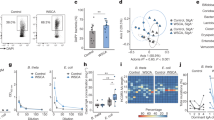
To characterize the heterogeneity of colon-resident MAIT cells, we profiled normal colon biopsies and matching blood samples from six individuals using multicolor flow cytometry. Each sample was analyzed with a panel of 13 monoclonal antibodies including those specific for activation and immunomodulatory markers (e.g., CD137, CD69, CD25, HLA-DR, PD-1, LAG-3, CTLA4, TIGIT, CD38, and CD244, see Supplementary Table 1 online). t-SNE analysis revealed that MR1-5-OP-RU-tetramer-positive MAIT cell populations in biopsies clustered only in three main regions. MAIT cells in these regions were rare in the blood indicating important phenotypic differences between mucosal and circulating MAIT cells (Fig. 1a and Supplementary Figure 1).
Intestinal MAIT cells are phenotypically homogeneous and cluster differentially from MAIT cells in blood. a t-SNE plots of flow cytometry-analyzed gut (red) and blood (blue) CD3+CD161+MR1-5-OP-RU tetramer+ MAIT cells. t-SNE analysis was performed after down-sampling and concatenation of all samples (paired blood and gut from six healthy donors). b t-SNE plots as in a showing relative expression of the indicated markers according to the color scale. c Percentage of MAIT cells expressing the indicated surface markers in gut vs. blood (n = 6). Horizontal bars represent mean ± sd. *P < 0.05 by Wilcoxon-rank test
Gut-resident MAIT cells differed from blood MAIT cells according to the expression patterns of activation and immunomodulatory markers (Fig. 1b). A larger number of intestinal MAIT cells showed expression of CD69, HLA-DR, CD25, TIGIT, PD-1, CTLA4, and LAG-3 as compared with blood-derived MAIT cells (Fig. 1c and Supplementary Figure 1).
Next, we analyzed the expression of transcription factors T-bet and RORγt, together with those of granzyme B, IFNγ, IL-17, IL-22, and TNFα after PMA stimulation of paired gut-resident and circulating MAIT cells from seven donors (Fig. 2 and Supplementary Table 2). Gut-resident MAIT cells distributed into distinct subpopulations, which minimally overlapped with blood-derived MAIT cells (Fig. 2a), indicating that, like the activation markers, the expression of cytokines and transcription factors was also distributed differently compared to circulating cells. About 9% of gut-resident MAIT cells expressed RORγt, and 65% were T-bet-positive, while in blood they were 1.5% and 40%, respectively. The percentage of gut-resident MAIT cells positive for all tested cytokines and granzyme B was significantly increased compared to that of blood (Fig. 2c). Unsupervised cluster analysis using DBSCAN identified 25 populations of MAIT cells (Fig. 2d). Populations 11, 12, 14, 20, and 25 contained higher frequencies of gut-derived MAIT cells (Supplementary Figure 2), whereas populations 2, 18, 19, 21, and 24 were more abundant in blood-derived MAIT cells. Populations 11, 12, and 14 in the gut showed high production of IL-17 and TNFα (Fig. 2e), while populations 11 and 12 also produced IFNγ. Population 20 mostly produced TNFα, while population 25 produced both TNFα and IFNγ. The populations significantly more abundant in the blood (2, 18, 19, 21, and 24, Supplementary Figure 2) instead produced very low amounts or no IFNγ, TNFα, IL-17, and IL-22 (Fig. 2e). These findings suggest that in the colon, MAIT cells express activation markers (Fig. 1) and are primed for production of pro-inflammatory cytokines in large amounts.
Intestinal MAIT cells are functionally homogeneous and cluster differentially from MAIT cells in blood. a t-SNE plots of flow cytometry-analyzed gut (red) and blood (blue) CD3+CD161+MR1-5-OP-RU tetramer+ MAIT cells. t-SNE analysis was performed after down-sampling and concatenation of all samples (paired blood and gut from seven healthy donors). b t-SNE plots as in a showing relative expression of the indicated markers according to the color scale. c Percentage of MAIT cells expressing the indicated intracellular marker in gut vs. blood (n = 7). Horizontal bars represent mean ± sd. d Semi-automated clustering using DBSCAN to identify MAIT cell populations (1–25) and e heatmap of their relative expression of the indicated markers in individual MAIT cell populations in gut vs. blood (n = 7). *P < 0.05 by Wilcoxon-rank test. The red asterisks denote populations significantly increased in gut vs. blood, the blue ones those decreased
These phenotypes and functions probably reflect constant stimulation by metabolites derived from commensal and opportunistic microflora. The expression of inhibitory molecules might provide a suitable mechanism to balance local continuous antigen stimulation. These observations suggest that the microflora and probably the gut microenvironment have important effects on tissue resident MAIT cells.
Potency hierarchy of MAIT cell-stimulatory metabolites
We then sought to determine whether the bacterial metabolites, which stimulate MAIT cells with different potencies,28 have different effects on MAIT cells. We used synthetic 5-OE-RU, 5-OP-RU, and RL-6,7-diMe presented by engineered A375 cells lacking functional MHC class I and class I-like molecules, transduced with a hybrid β2M-human MR1 gene (A375.MR1) (ref. 29 and Supplementary Figure 3). These APCs were engineered in order to limit alloreactive stimulation and prevent the effects of MHC class-I-binding inhibitory molecules potentially expressed by MAIT cells.
All three synthetic MAIT cell antigens increased MR1 surface expression (Supplementary Figure 3a), thereby confirming the stabilization capacity of these molecules.30 In this assay, RL-6,7-diMe induced MR1 upregulation less efficiently than the other compounds (Supplementary Figure 3b). At high concentrations (50–500 µM), 5-OP-RU was more effective than the other compounds (Supplementary Figure 3b).
To examine the potencies of the three synthetic antigens in stimulating MAIT cells, we selected two human MR1-restricted MAIT cell clones, MRC25 and SMC3, which express canonical TRAV1.2–TRAJ33 TCRα chains paired to different TCRβ chains (Table 1) and respond to E. coli-infected APCs.2,29 The release of IFNγ by MAIT cell clones upon antigen recognition was used to measure the potency (effective concentration for half-maximum response, EC50) of each tested compound. 5-OP-RU and 5-OE-RU were highly stimulatory to both clones, whereas RL-6,7-diMe displayed lower potency (Fig. 3a, b). SMC3 was slightly more sensitive than MRC25 (Fig. 3c) and each compound showed comparable EC50 values in each of the two clones (Fig. 3c and Table 1). The stimulatory capacity of these antigens displayed a consistent potency hierarchy of 5-OP-RU > 5-OE-RU > RL-6,7-diMe.
Bacterial riboflavin-related synthetic antigens activate MAIT cells in a dose-dependent manner. a, b Dose-dependent IFNγ response of the MAIT cell clone (a) SMC3 and (b) MRC25 after stimulation with A375.MR1 cells and RL-6,7-diMe (diamonds), 5-OP-RU (circles), 5-OE-RU (squares), or in the absence of antigen (ns, not stimulated). c Log10EC50 values of RL-6,7-diMe (diamonds), 5-OE-RU (squares), and 5-OP-RU (circles) inducing half maximum IFNγ release by MAIT cell clones as in a, b. Data show mean ± sd of duplicates and are representative of two independent experiments. d Flow cytometry assessing TCR downregulation on MAIT cells (CD3+Vα7.2+CD161hi) present in PBMCs of a representative donor after co-culture with A375.MR1 and RL-6,7-diMe (50 µM), 5-OE-RU (0.5 µM), 5-OP-RU (5 nM) or E. coli extract (5 × 107 CFU/mL). e, f Median fluorescence intensity (MFI) of the TCR on MAIT cells (Vα7.2+CD161hi as in d of another donor in response to varying doses of e 5-OP-RU (circles), 5-OE-RU (squares), RL-6,7-diMe (diamonds) or f E. coli extract. One representative out of five experiments is shown
We next compared the three synthetic antigens and E. coli cell extract for their capacity to stimulate MAIT cells present in freshly isolated PBMC. Due to the TCR heterogeneity of circulating MAIT cells, the antigen-induced downregulation of the TCR in polyclonal MAIT cell populations was used to determine the EC50 of each compound; one such example is shown in Fig. 3d. Dose response curves confirmed the hierarchy of potencies observed with MAIT clones, (Fig. 3e, f and Table 1). Despite inter-donor variability, 5-OP-RU at EC50 doses induced upregulation of CD69 in stimulated circulating MAIT cells from five healthy donors, and CD25 and CD137 in three out of five donors (Supplementary Figure 4). In contrast, EC50 doses of 5-OE-RU and RL-6,7-diMe did not induce comparable MAIT cell activation in the majority of donors (Supplementary Figure 4). These findings are in line with other studies conducted with T cells engineered with TCR genes from one MAIT clone and indicating 5-OP-RU as a potent antigen.16,18
We next assessed whether the same hierarchy was observed in the presence of APCs expressing physiological levels of MR1 as those present in blood. PBMCs from healthy individuals were exposed for a short time to synthetic antigens without the addition of exogenous APCs. The downregulation of the TCR and concomitant upregulation of the activation markers CD69, CD25, and CD137 on MAIT cells was analyzed by flow cytometry (Supplementary Figure 5a,b). Also under these experimental conditions, the previously observed potency hierarchy was observed. However, the efficiency of stimulation was much lower; the EC50 concentrations calculated from TCR downregulation were ~102–10 3 times higher than those observed in the presence of MR1-transfected APCs (EC50 5-OP-RU, 43.2 nM; EC50 5-OE-RU, 3.21 µM), and even at high concentrations (500 µM) of RL-6,7-diMe, TCR downregulation was barely detectable.
Phenotypic changes of MAIT cells after activation by synthetic antigens
To analyze the antigen-induced phenotypic changes, circulating MAIT cells were stimulated for 24 h with A375.MR1 cells and three doses each of the three synthetic antigens and two concentrations of E. coli extract (Fig. 4). The activation and immunomodulatory markers that were investigated are described in Supplementary Table 1.
Multiple populations of ex vivo MAIT cells respond to riboflavin-related antigens. MAIT cell response of one healthy donor after PBMC stimulation for 24 h with A375.MR1 cells and a–c RL-6,7-diMe, d–f 5-OE-RU, g–i 5-OP-RU, j, k E. coli extract or l medium (unstimulated). The response to EC50 and ED50 doses of each antigen are presented in the middle panels (b, e, h, k). t-SNE analysis of CD3+Vα7.2+CD161hi MAIT cells after down-sampling and concatenation of all conditions. Warm color regions of the t-SNE map depict regions of higher cell density. Experiments were performed twice with comparable results in two different donors
Unstimulated MAIT cells (Fig. 4l) and those stimulated by RL-6,7-diMe clustered in two major overlapping areas (Fig. 4a–c). RL-6,7-diMe was active only at the highest dose tested, confirming the weak antigenicity of this metabolite. At low concentrations, 5-OE-RU and 5-OP-RU induced minor but clear changes in cluster distribution when compared to unstimulated MAIT cells (Fig. 4d, g vs. l). At higher doses, both 5-OE-RU and 5-OP-RU induced the appearance of novel clusters (Fig. 4e, f, h, i), distinct from those observed after stimulation with RL-6,7-diMe. The main cluster of resting cells observed in the unstimulated condition (Fig. 4l) disappeared after stimulation with 50 nM 5-OE-RU and with 5 nM 5-OP-RU, confirming the differential potency of these two antigens.
The unpurified antigens present in E. coli extracts induced a dose-dependent response with the appearance of clusters very similar to those induced by 5-OE-RU and 5-OP-RU (Fig. 4j, k vs. e, f and h, i).
When circulating MAIT cells were stimulated for 72 h with A375.MR1 cells and the EC50 of each antigen, the expression of each marker was different from that observed after 24 h (Supplementary Figure 6), indicating that continuous stimulation induces further phenotypic changes.
These data indicated that freshly isolated MAIT cells respond to synthetic antigens by differential modulation of surface markers involved in T-cell activation and regulation.
MAIT cells split into different groups upon stimulation with different antigens
To further investigate the differential MAIT cell response to weak (RL-6,7-diMe), intermediate (5-OE-RU), and strong (5-OP-RU) antigens, PBMCs were stimulated with A375.MR1 and with each of these synthetic analogs at concentrations inducing TCR downregulation. The analysis of flow-cytometry data was performed at two time-points (24 and 72 h after stimulation) using FlowSOM, a semi-supervised clustering and dimensional reduction algorithm.31 MAIT cells were fit to the best matching node according to the expression of surface markers. Nodes 5, 8, 11, 17, 19, 20, 21, 22, 23, 24, and 25 are shown in Fig. 5a, while all the other nodes are shown in Supplementary Figure 7a. As illustrated by the node size, the majority of MAIT cells stimulated with RL-6,7-diMe for 24 h distributed in the self-organizing map similarly to unstimulated cells, and expressed low levels or none of the investigated markers (Fig. 5b, node 5). In contrast, cells stimulated by 5-OE-RU or 5-OP-RU were fit to three similar nodes (20, 23, and 25) with different phenotypes (Fig. 5b). Nodes 23 and 25 contained MAIT cells expressing the activation markers CD137, CD69, CD25, and the regulatory molecules PD-1 and LAG-3 (Fig. 5b). Cells in nodes 23 and 25 were different because they expressed different levels of investigated marker. For instance, MAIT cells in node 25, but not in node 23, displayed low-surface expression of CD3, indicating a strong response to antigen of these cells (Fig. 5b).
Clusters of ex vivo-stimulated MAIT cells according to their phenotypic profile. FlowSOM analysis of MAIT cells from one healthy donor after PBMC stimulation with A375.MR1 cells and RL-6,7-diMe, 5-OE-RU, 5-OP-RU, (EC50 concentrations), and E. coli extract (107 CFU/mL, ED50), or medium (unstimulated) for 24 h and 72 h. a Minimal spanning tree illustrating 25 nodes containing MAIT cells stimulated with various doses of antigens and E. coli extract. Radar charts show expression of markers defining each node. b, c Expression of indicated surface markers on unstimulated and antigen stimulated MAIT cells after b 24 and c 72 h in the main node is shown. Minor nodes are shown in Supplementary Fig. 6. Numbers indicate main nodes defined by the semi-supervised FlowSOM algorithm in a. Pie charts indicate the median fluorescence intensity of markers and node size corresponds to the number of MAIT cells in each population. Experiments were performed twice with comparable results in three healthy donors
MAIT cells activated by E. coli extracts were more similar to that induced by 5-OE-RU than to the activation observed in response to RL-6,7-diMe (Fig. 5b and Supplementary Figure 7b). This analysis revealed MAIT cell responsiveness can be classified into discrete steps, uniquely depicting their response to weak, intermediate, and strong antigens.
MAIT cells analyzed 72 h after stimulation did not show down-modulation of Vα7.2 TCR chain and CD3 (Fig. 5c and Supplementary Figure 7c), independently of the stimulating antigen, confirming the transient nature of CD3–TCR complex downregulation following antigen recognition.32
Cells responsive to the weak antigen RL-6,7-diMe were dispersed across nodes 9–11. MAIT cells in node 9 and 10 expressed PD-1, and those in node 9 co-expressed CD38 and low levels of LAG-3 and HLA-DR (Supplementary Figure 7c). Thus, the analysis of MAIT cells 72 h after stimulation with the weak RL-6,7-diMe antigen clearly displayed differences to unstimulated cells.
After 72 h, MAIT cells stimulated with 5-OE-RU, 5-OE-RU, and E. coli extract fit to nodes 22 and 24, and expressed the activation markers CD25, CD69, and CD38, together with the immunomodulatory molecules LAG-3 and PD-1 (Fig. 5c). Cells in node 24 also expressed two additional activation markers; HLA-DR and CD137, thus indicating a higher level of activation in this MAIT cell population than that of node 22 (Fig. 5c). Collectively, these data suggest that the potency of MR1-ligands strongly influences MAIT cell expression of activation and immunomodulatory markers whose differential combinations distinguish responsive MAIT cell populations.
Bacterial cell culture conditions modulate MAIT cell-stimulating capacity
Bacterial growth conditions exert important effects on microbial metabolism and intestinal environmental conditions shape the development and composition of the adaptive immunity.33 According to this paradigm, we investigated whether E. coli adapted to grow in gut-related conditions acquire unique capacities to induce MAIT cell activation and phenotypic changes.
Bacterial metabolites were extracted from E. coli grown in different conditions including altered carbon sources and concentrations, various oxygen tensions, or various starting pH values. Bacterial extracts, normalized for their optical density and expressed as CFU/mL were assessed for the capacity to induce IFNγ release by MAIT cell clones. By comparing the half-maximal effective dose response (ED50) to bacterial extract, we assessed the antigenic potency of E. coli harvested during exponential or stationary phase, when grown in nutrient-rich LB medium or in minimal M9 medium. The ED50 stimulating SMC3 cells was significantly lower with extracts from E. coli harvested in stationary vs. exponential phase (Fig. 6a, b), suggesting accumulating antigens are a product of secondary metabolism, active during stationary phase growth. The rich nutrient composition of LB medium also favored stimulation and the ED50 calculated for LB and M9 were 105.18 ± 0.16 and 107.44 ± 0.09 (mean ± sd), respectively, for cells harvested during their exponential growth, and 104.51 ± 0.07 and 105.93 ± 0.06, respectively, for cells grown at stationary phase (Fig. 6a, b). When stationary phase E. coli were grown in M9 medium with a starting pH ranging from 6.0 to 7.5 (the physiological pH range within the distal intestinal tract34), all extracts had similar potency (Fig. 6c), indicating that tested pH conditions do not significantly modulate the bacterial stimulatory ability.
Bacterial culture conditions influence the response of MAIT cells. Response of the MAIT cell clone SMC3 to A375.MR1 cells in the presence of titrated extracts from E. coli grown in exponential (open circles) and stationary (closed circles) phases in a complete LB medium and b synthetic M9 medium. c, d Response of SMC3 cells to A375.MR1 stimulated with titrated extracts from E. coli grown at stationary phase in c synthetic medium with different starting pH and d LB medium in aerobic (closed circles) and anaerobic (open circles) conditions. a–d Results are expressed as mean ± sd of IFNγ release of triplicate cultures. e, f ED50 values of E. coli extracts inducing IFNγ release by SMC3 cells in response to A375.MR1 cells incubated with extracts of E. coli grown at stationary phase in M9 medium supplemented with 40 mM of the indicated sole carbon sources or with 4 mM glucose. Log10 ED50 values were calculated based on the response to titrating amounts of bacterial extracts equivalent to 103–109 CFU/mL. Results are expressed as mean + sd of IFNγ release of triplicate cultures. *P ≤ 0.05, ****P ≤ 0.0001, analyzed using one-way ANOVA with Dunnett’s multiple comparison test against values obtained with standard 40 mM glucose. Data show triplicates measured as technical replicas and are representative of two independent experiments
To test whether oxygen modulates the production and accumulation of MAIT antigens, we compared the stimulatory capacity of bacterial extracts obtained from E. coli grown under aerobic or anaerobic conditions. Extracts from anaerobically grown stationary phase E. coli were more stimulatory than those from aerobically grown E. coli (Fig. 6d), suggesting that oxygen limitation enhances the capacity of E. coli to activate MAIT cells.
Finally, we assessed the impact of carbon sources during stationary growth of E. coli on their MAIT-stimulatory capacity. As indicated by low ED50 values, extracts from E. coli grown with 40 mM glucose were more potent than those from E. coli grown with 4 mM glucose (Fig. 6e and Supplementary Figure 8a). Supplementing M9 medium with a variety of sole carbon sources at 40 mM and comparing the ED50 obtained with the same concentration of glucose, we observed that extracts from E. coli grown with xylose showed a comparable ED50, while extracts from bacteria grown with ribose were more potent (Fig. 6e and Supplementary Figure 8b,c). In contrast, extracts from E. coli grown with sodium pyruvate showed reduced stimulatory capacity (Fig. 6e and Supplementary Figure 8d). When non-fermentable carbon sources like glycerol, lactate and acetate were added as sole carbon source, E. coli extracts were less efficient than extracts from bacteria grown in 40 mM glucose (Fig. 6f and Supplementary Figure 8e-g). Taken together these results indicate that growth phase, oxygen tension and carbon sources, but not pH, influence MAIT cell antigenicity of E. coli lysates.
Accumulation of stimulatory metabolites in different bacterial growth conditions
To gain insight into the bacterial physiology that underlies the observed culture-dependent alteration of MAIT cell stimulation, we performed untargeted metabolome analysis of E. coli grown in defined media with seven different sole carbon sources (glucose, xylose, ribose, pyruvate, glycerol, lactate, and acetate) and at four different pH values (6.0, 6.5, 7.0, and 7.5) for glucose. The identical bacterial lysates were also used to stimulate MAIT cells and determine their ED50. To assess how the 573 detected metabolites changed between growth conditions, we performed a pair-wise comparison (t-test) using glucose (pH 7) as the reference condition (Supplementary Table 3). To relate metabolite changes to actual pathways, we performed a metabolite set enrichment analysis on significantly altered metabolites (log2 fold change) using the pathway definitions in the KEGG database.35,36 E. coli grown in ribose led to the strongest stimulation amongst all conditions (Fig. 6e), coinciding with metabolites enriched in purine and folate metabolism (Supplementary Figure 9a). In contrast, pyruvate and lactate conditions that poorly stimulated MAIT cells (Fig. 6e, f) showed reduced abundance of purine metabolites, illustrated by pathway enrichment of decreasing metabolites (Supplementary Figure 9b). Both purine and folate pathways are directly connected to the riboflavin pathway20 and are thus potentially linked to the generation of MAIT cell antigens.16,18 Hence, these findings support our hypothesis that altered bacterial growth conditions affect the production of riboflavin precursors and correlate with the altered capacity of bacteria to stimulate MAIT cells.
To further investigate this possibility, we focused on the abundance of metabolites in the riboflavin pathway. Compared to glucose as a sole carbon source, E. coli grown with ribose, xylose, and glycerol showed an increase in abundance of several riboflavin precursors including; GTP, the first metabolite in the pathway; 2,5-diamino-6-(5-phospho-d-ribosylamino)pyrimidin-4(3H)-one; 5-amino-6-(5′-phospho-d-ribitylamino)uracil; 5-A-RU, and RL-6,7-diMe (Fig. 7). In contrast, the levels of these metabolites were unchanged or reduced in the weakly stimulating acetate, lactate or pyruvate conditions (Fig. 7). These findings consolidated our hypothesis that E. coli growth conditions facilitating MAIT cell stimulation coincide with an increased abundance of riboflavin biosynthesis intermediates.
Accumulation of riboflavin and precursor metabolites in E. coli depending on carbon source. Log2 fold change in the abundance of riboflavin-precursor metabolites in E. coli extracts grown in M9 medium supplemented by the indicated sole carbon sources (40 mM) or glucose at 4 mM, relative to that obtained with M9 supplemented by 40 mM glucose (n = 4). Metabolite amounts were determined by mass spectrometry. Log2 fold changes of metabolites are color coded
Next, we analyzed the levels of the riboflavin-derived antigens 5-OE-RU, 5-OP-RU, RL-6,7-diMe, and their precursor 5-A-RU. Compared to glucose at 40 mM, 5-A-RU was significantly increased in xylose and glycerol (Fig. 8a), RL-6,7-diMe was increased in xylose (Fig. 8b), and 5-OE-RU in 4 mM glucose (Fig. 8c). The potent MAIT cell stimulator 5-OP-RU was increased in xylose and ribose conditions (Fig. 8d). Taken together, these results show hexose, pentose and glycerol conditions, but not pyruvate, lactate, and acetate conditions increased production of MAIT antigens and the abundance of 5-A-RU. In agreement with the unchanged MAIT cell-stimulating capacity of bacterial extracts derived from pH-adjusted bacterial cultures, the abundance of riboflavin-related antigens was also stable (Supplementary Figure 10a-d). Significant correlations between the abundance of these metabolites and the ED50 of the corresponding extracts were observed for all investigated molecules (Fig. 8e, f, h), except 5-OE-RU (Fig. 8g). Overall, these findings suggest a link between accumulation of riboflavin-related metabolites and the potential of bacteria to stimulate MAIT cells.
Abundance of riboflavin-precursor antigens in E. coli is modulated by carbon sources and their concentration. a–d Relative abundance expressed as arbitrary units (AU) of indicated pyrimidines and ribityllumazines in extracts of E. coli grown in M9 medium supplemented by the indicated sole carbon sources. e–h Correlation of the metabolite levels with the E. coli Log10 ED50 of MAIT cell stimulation (as in Fig. 5e, f). a, e 5-A-RU (m/z 275.1004), b, f RL-6,7-diMe (m/z 325.1155), c, g 5-OE-RU (m/z 315.0949), and d, h 5-OP-RU (m/z 329.1105). In this type of analysis, 5-OP-RU cannot not be distinguished from reduced 6-hydroxymethyl-8-d-ribityllumazine (rRL-6-CH2OH,) as they have identical masses and fragment spectra.16,18,19 *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.001, analyzed using Kruskal–Wallis test with Dunnett’s multiple comparison test against values obtained with standard 40 mM glucose. Data show triplicates measured as technical replicas and are representative of two independent experiments
In conclusion, the abundance of MAIT cell-stimulating metabolites significantly varies in E. coli depending on growth conditions and by extension on the local host-environment. Growth with hexose, pentose or glycerol maximized the presence of such antigenic metabolites, whereas acetate, lactate, and pyruvate conditions reduced their abundances. Thus, adaptation of the microflora to gut-related physiological growth conditions impacts on the abundance and type of stimulatory metabolites, which in turn may modulate MAIT cell activation and heterogeneity.
MAIT cells sense bacterial growth conditions
The correlation observed between accumulation of MAIT cell antigenic metabolites depending on E. coli growth conditions and capacity to stimulate MAIT cells prompted us to investigate the phenotype of MAIT cells responding to bacteria exposed to diverse gut-related growth conditions.
Flow cytometric analysis of blood MAIT cells stimulated with E. coli extracts from exponential phase revealed low levels expression of CD69 and PD-1 (Fig. 9), while those stimulated with stationary phase E. coli showed high CD69, PD-1, CD25, and LAG-3 expression (Fig. 9 and Supplementary Figure 11). Growth of E. coli under anaerobic conditions also led to significant activation of freshly isolated MAIT cells (Fig. 9 and Supplementary Figure 11), confirming the data obtained with stimulation of a MAIT clone (Fig. 6). In contrast, bacteria grown in aerobic conditions or harvested during exponential growth phase induced weaker expression of these activation markers resulting in a t-SNE map more similar to the unstimulated control (Fig. 9 and Supplementary Figure 11).
Ex vivo MAIT cell responses are affected by E. coli growth conditions. t-SNE analysis performed on CD3+Vα7.2+CD161hi MAIT cells after PBMC stimulation in co-culture with A375.MR1 and E. coli extracts from the indicated growth conditions. Extracts were normalized to their OD600 (as in Fig. 5b, d). MAIT cells from one healthy donor were analyzed after down-sampling and concatenation of all conditions. Colors indicate cell expression levels of CD69 (upper panels) and PD-1 (lower panels). Experiments were performed twice with comparable results in two different donors
In conclusion, bacterial growth conditions directly modulate the accumulation of MAIT cell-stimulating metabolites and indirectly control their activation and expression of inhibitory receptors. This mechanism allows a fine regulation of MAIT cell response in the gut.
Discussion
The human gut has been defined as an anaerobic bioreactor, which maintains a local flora composed of a few divisions of bacteria37 and with slow microbial growth dynamics.38 The metabolic repertoire of gut microorganisms is a key factor in shaping the phenotype, composition, and functions of resident immune cells.39 These effects are mediated through the release of small metabolites that influence a cascade of metabolic events within host cells.40 Some bacterial metabolites serve as antigens presented by MR1 and stimulating MAIT cells.16,18,19,41 The contribution of bacterial metabolites produced in tissues to MAIT cell activation and phenotypic differentiation remains poorly investigated.
Our studies showed that MAIT cells in human intestinal mucosa are distributed in a few distinct populations, which represent a minority of cells in the circulating pool. MAIT cells in gut biopsies expressed different combinations of HLA-DR, CD25, CD69, and CD137, suggesting variable states of activation. In particular, about 50% of MAIT cells were CD137+, a marker of recently activated T cells.42 This finding is in accordance with the known capacity of intestinal bacterial flora to stimulate immune cells.43,44 Some MAIT cell populations expressed inhibitory receptors, including TIGIT, PD-1, CTLA4, and LAG-3. About 50% of MAIT cells in the gut were TIGIT+ or PD-1+; however, only a minor percentage of cells co-expressed these two markers, while most of TIGIT+ cells were LAG-3- and CTLA4-negative. These findings indicate that active stimulation occurs in the gut and that MAIT cells rely on different inhibitory mechanisms to dampen their responsiveness to antigen. Most of the MAIT cells observed in colon biopsies express T-bet and only some RORγt transcription factors distinctive of TH1 and TH17 functional phenotypes, respectively. Accordingly, we found that they produce large amounts of pro-inflammatory cytokines and granzyme B when compared to circulating MAIT cells. The expression of activation markers together with that of inhibitory molecules and the efficient cytokine production suggests that MAIT cells in the colon are primed T cells that might either (i) become readily reactivated, (those not expressing inhibitory receptors) or (ii) remain non-responsive after antigen stimulation (being regulated by inhibitory receptors). The proper balance between these cell populations might be relevant to local immunity.
Notably, a large number of MAIT cells responded to extracts from E. coli grown under conditions simulating key features of the colon microenvironment (carbon sources, oxygen tension, and growth phase) by expressing TIGIT or PD-1, while a few cells expressed LAG-3 and CTLA4. Although similar phenotypes were observed in MAIT cells from gut biopsies, it is not possible to conclude that the observed ex vivo phenotypes is a consequence of the same stimulation applied in vitro.
In agreement with published work,16,18,22 the activity of 5-OP-RU was very potent as low picomolar concentrations induced IFNγ release in MAIT cell clones and TCR downregulation in freshly isolated polyclonal MAIT cells. In contrast, 5-OE-RU and RL-6,7-diMe, respectively, were 102–103 and 5 × 105–109 fold less potent than 5-OP-RU in TCR downregulation. The same potency hierarchy was observed with APC displaying low physiological levels of MR1, although TCR down modulation and high expression of CD25, CD69, and CD137 markers required higher antigen doses. Using such APC, RL-6,7-diMe was poorly stimulatory, even at high doses, thus implicating the importance of MR1 protein levels on the surface of APC in defining the contribution of each antigen to activation.
Stimulation with 5-OP-RU and 5-OE-RU also induced the expression of surface markers found in ex vivo analyzed gut-derived MAIT cells. The weak RL-6,7-diMe antigen did not induce the same upregulation of activation and inhibitory markers at either high doses or after prolonged stimulation. Similar observations were found with the FlowSOM self-organizing map algorithm. This analysis also showed that the investigated markers appeared on stimulated cells in discrete steps, characterized by early appearance of activation markers followed by the inhibitory ones. At a 72 h time point, most cells grouped in two major nodes after stimulation with 5-OP-RU and 5-OE-RU and E. coli, but not with RL-6,7-diMe. Thus, different antigens induce different phenotypic changes in MAIT cells.
Having characterized the stimulating capacity of each antigen, we compared their levels in bacteria grown under different conditions. Bacterial metabolism is influenced by environmental cues including nutrient and metabolite concentrations, pH, oxygen tension, as well as microbial cell replication time, and host immune responses.23,24,25,26,27 By altering in vitro culture conditions we identified several factors, also present in the gut, capable of modulating the capacity of E. coli to activate MAIT cells. E. coli grown in stationary phase and slowly replicating, as occurs in human gut,38 stimulated MAIT cells more efficiently than bacteria grown in nutrient-rich media and in exponential phase. Moreover, the absence or low tension of oxygen during bacterial growth enhanced MAIT cell stimulation. Probably, stationary phase and anaerobiosis enhance MAIT cell stimulation by multiple mechanisms with accumulation of MAIT stimulating metabolites providing a critical contribution. In addition, the possibility exists that the relative abundance of non-stimulatory MR1-binding metabolites might affect stimulation.
Metabolomics analysis of extracts from bacteria grown with a sole carbon source indicated a correlation between the increase of purine, vitamin B9 (folic acid) and B2 (riboflavin) pathways, and stimulatory capacity. Metabolites from the three pathways were abundant in E. coli grown in defined media supplemented with ribose, which induced the strongest stimulation of MAIT cells. Furthermore, metabolites of the riboflavin pathway showed high levels in the ribose-supplemented condition, and a significant correlation was observed between low EC50 (high potency) and increased levels of 5-OP-RU, RL-6,7-diMe, and its precursor 5-A-RU.
Extracts of E. coli grown with low oxygen and in stationary phase also induced expression of surface markers on MAIT cells resembling the markers present on ex vivo investigated colon MAIT cells. These similarities indicate that stimulation occurs inside the gut, possibly facilitated by the local accumulation of potent microbial antigens.
Open issues are: (i) how the steep oxygen gradients are in the gut45 and (ii) how the precise localization of bacteria in the colon directly affects MAIT cell stimulation. The half-lives of 5-OP-RU and 5-A-RU are very short,22 and when produced by bacteria located in the central part of the gut lumen they are unlikely to stimulate MAIT cells located in intraepithelial space and lamina propria as they must pass through the glycocalyx that forms a thick bi-layered structure in the colon.33 In contrast, 5-OP-RU and 5-A-RU released by bacteria growing in the microaereophilic zone adjacent to epithelial cells are more available for presentation by MR1. Since the latter bacteria are directly affected by local immune responses, a reciprocal influence can be envisaged. Therefore, the balance between local microbial surveillance and bacterial riboflavin metabolism might be relevant to MAIT cell activation.
In conclusion, these studies showed that different populations of MAIT cells in the gut exhibit phenotypes of cells with recent antigen experience either ready for further responses or regulated by inhibitory receptors. Similar phenotypes are induced by metabolite antigens that accumulate under low oxygen conditions and during slow bacterial growth. Thus, MAIT cells represent an innate-like population of T cells with the capacity to sense the metabolic state of enteric bacteria.
Methods
Human samples and cell lines
Colon biopsies and paired blood samples were obtained from the Gastroenterology Unit, University Hospital Basel, Switzerland. Samples were from patients under colon cancer screening and were normal at histological evaluation. The study was approved by the Swiss ethics commission (no. 139-13) and written informed consent was obtained from all subjects. For further details and description of T-cell isolation from biopsies, and used cell lines, see Supplementary Methods.
Functional assays
For description of T-cell stimulation, flow cytometry, multidimensional analyses, metabolite synthesis, extraction of bacterial metabolites, metabolomics methods, and statistical analyses, see Supplementary Methods.
References
Treiner, E. et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003).
Lepore, M. et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRbeta repertoire. Nat. Commun. 5, 3866 (2014).
Walker, L. J. et al. Human MAIT and CD8alphaalpha cells develop from a pool of type-17 precommitted CD8+T cells. Blood 119, 422–433 (2012).
Reantragoon, R. et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210, 2305–2320 (2013).
Gold, M. C. et al. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J. Exp. Med. 211, 1601–1610 (2014).
Dias, J., Leeansyah, E. & Sandberg, J. K. Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines. Proc. Natl Acad. Sci. USA 114, E5434–E5443 (2017).
Dusseaux, M. et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117, 1250–1259 (2011).
Tang, X. Z. et al. IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. J. Immunol. 190, 3142–3152 (2013).
Seach, N. et al. Double-positive thymocytes select mucosal-associated invariant T cells. J. Immunol. 191, 6002–6009 (2013).
Koay, H. F. et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat. Immunol. 17, 1300–1311 (2016).
Gold, M. C. et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 8, e1000407 (2010).
Le Bourhis, L. et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11, 701–708 (2010).
Chen Z., et al. Mucosal-associated invariant T-cell activation and accumulation after in vivo infection depends on microbial riboflavin synthesis and co-stimulatory signals. Mucosal Immunol. 10, 58–68 (2016).
Ussher, J. E. et al. CD161++CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur. J. Immunol. 44, 195–203 (2014).
Jo, J. et al. Toll-like receptor 8 agonist and bacteria trigger potent activation of innate immune cells in human liver. PLoS Pathog. 10, e1004210 (2014).
Kjer-Nielsen, L. et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012).
Patel, O. et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 4, 2142 (2013).
Corbett, A. J. et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 (2014).
Lopez-Sagaseta, J. et al. MAIT recognition of a stimulatory bacterial antigen bound to MR1. J. Immunol. 191, 5268–5277 (2013).
Bacher, A., Eberhardt, S., Fischer, M., Kis, K. & Richter, G. Biosynthesis of vitaminb2 (riboflavin). Annu. Rev. Nutr. 20, 153–167 (2000).
McWilliam, H. E. et al. The intracellular pathway for the presentation of vitamin B-related antigens by the antigen-presenting molecule MR1. Nat. Immunol. 17, 531–537 (2016).
Mak, J. Y. et al. Stabilizing short-lived Schiff base derivatives of 5-aminouracils that activate mucosal-associated invariant T cells. Nat. Commun. 8, 14599 (2017).
Louis, P., Scott, K. P., Duncan, S. H. & Flint, H. J. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 102, 1197–1208 (2007).
Spor, A., Koren, O. & Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279–290 (2011).
Guarner, F. & Malagelada, J. R. Gut flora in health and disease. Lancet 361, 512–519 (2003).
Ley, R. E., Peterson, D. A. & Gordon, J. I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 (2006).
Hooper, L. V. & Macpherson, A. J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10, 159–169 (2010).
Birkinshaw, R. W., Kjer-Nielsen, L., Eckle, S. B., McCluskey, J. & Rossjohn, J. MAITs, MR1 and vitamin B metabolites. Curr. Opin. Immunol. 26, 7–13 (2014).
Lepore, M., et al. Functionally diverse human T cells recognize non-microbial antigens presented by MR1. Elife 6, e24476 (2017).
Mondot, S., Boudinot, P. & Lantz, O. MAIT, MR1, microbes and riboflavin: a paradigm for the co-evolution of invariant TCRs and restricting MHCI-like molecules? Immunogenetics 68, 537–548 (2016).
Young, M. H. et al. MAIT cell recognition of MR1 on bacterially infected and uninfected cells. PLoS ONE 8, e53789 (2013).
Liu, H., Rhodes, M., Wiest, D. L. & Vignali, D. A. On the dynamics of TCR:CD3 complex cell surface expression and downmodulation. Immunity 13, 665–675 (2000).
Agace, W. W. & McCoy, K. D. Regionalized development and maintenance of the intestinal adaptive immune landscape. Immunity 46, 532–548 (2017).
Fallingborg, J. Intraluminal pH of the human gastrointestinal tract. Dan. Med. Bull. 46, 183–196 (1999).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A. & Gordon, J. I. Host-bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005).
Korem, T. et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349, 1101–1106 (2015).
Brestoff, J. R. & Artis, D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 14, 676–684 (2013).
Koh, A., De Vadder, F., Kovatcheva-Datchary, P. & Backhed, F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 (2016).
Soudais, C. et al. In vitro and in vivo analysis of the Gram-negative bacteria-derived riboflavin precursor derivatives activating mouse MAIT cells. J. Immunol. 194, 4641–4649 (2015).
Sanchez-Paulete, A. R. et al. Deciphering CD137 (4-1BB) signaling in T-cell costimulation for translation into successful cancer immunotherapy. Eur. J. Immunol. 46, 513–522 (2016).
Round, J. L. & Mazmanian, S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 (2009).
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011).
Espey, M. G. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic. Biol. Med. 55, 130–140 (2013).
Acknowledgements
We thank Shivani Koolarina, Zi-Xin Wong, Lena Angman, Sebastiano Sansano, Anouk Lehmann, Emmanuel Traunecker, and Danny Labes for technical support, Paula Cullen and Thérèse Resink for reading of the manuscript. The MR1-tetramer technology was developed jointly by James McCluskey, Jamie Rossjohn, and David Fairlie, and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne. This work was supported by the core funds of the Department of Biomedicine and SIgN and by the Swiss National Science Foundation (310030-149571 and -173240 to G.D.L.).
Author information
Authors and Affiliations
Contributions
M.S., L.M., and G.D.L. conceived and designed experiments. M.S., A.C., M.Z., J.S., and S.B. performed experiments and analyzed data. T.R. contributed to data analysis. M.L. and A.K. provided cells, P.H. provided clinical samples, A.E., F.C., and N.P. provided bacterial samples, E.A., M.P., and G.C. provided synthetic compounds. U.S. supervised metabolomics analysis, L.M. and G.D.L. supervised the work. M.S., L.M., and G.D.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Schmaler, M., Colone, A., Spagnuolo, J. et al. Modulation of bacterial metabolism by the microenvironment controls MAIT cell stimulation. Mucosal Immunol 11, 1060–1070 (2018). https://doi.org/10.1038/s41385-018-0020-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-018-0020-9
This article is cited by
-
MR1 antigen presentation to MAIT cells and other MR1-restricted T cells
Nature Reviews Immunology (2024)
-
MAIT cells and their implication in human oral diseases
Inflammation Research (2022)
-
MAIT cells, guardians of skin and mucosa?
Mucosal Immunology (2021)
-
Exhaustion in tumor-infiltrating Mucosal-Associated Invariant T (MAIT) cells from colon cancer patients
Cancer Immunology, Immunotherapy (2021)
-
Human unconventional T cells in Plasmodium falciparum infection
Seminars in Immunopathology (2020)