Abstract
P. gingivalis (Pg) is an oral pathogen with the ability to induce oral dysbiosis and periodontal disease. Nevertheless, the mechanisms by which mucosal responses to the oral microbiota in the presence of specific pathogens such as Pg could abrogate the host-microbe symbiotic relationship leading to periodontitis remain unclear. Herein, we identified the Notch-1/PLA2-IIA axis as a new molecular pathway through which Pg could be specifically modulating oral epithelial antimicrobial and inflammatory responses. Pg activated Notch-1, and inhibition or silencing of Notch-1 completely abrogated Pg-induced PLA2-IIA in oral epithelial cells (OECs). Activation of Notch-1 and PLA2-IIA production were associated with Pg-produced gingipains. Other oral Gram-positive and Gram-negative species failed to induce similar responses. Pg enhanced OEC antimicrobial activity through PLA2-IIA. Increased Notch-1 activation correlated with higher PLA2-IIA gingival expression and changes in the abundance of specific oral bacteria phyla during periodontal disease. Oral bacterial species exhibited differential antimicrobial susceptibility to PLA2-IIA. These findings support previous evidence suggesting an important role for epithelial Notch-1 activation and PLA2-IIA production during health and disease at mucosal surfaces, and provide new mechanistic information concerning the regulation of epithelial antimicrobial and pro-inflammatory responses modulated by oral pathogenic bacteria associated with periodontal disease.
Similar content being viewed by others
Introduction
Although specific oral pathogenic bacterial species such as Porphyromonas gingivalis (Pg) have been related to established periodontal lesions and can recapitulate periodontal disease in animal models, the early events associated with epithelial-oral pathogen interactions in a complex microbial ecology that creates a microenvironment enabling the initiation of this chronic inflammatory disease remain unclear.
Pg is an oral Gram-negative (G−), anaerobic bacterium that has been broadly associated with periodontal disease1 and systemic chronic inflammatory disorders such as atherosclerotic cardiovascular disease, diabetes, and rheumatoid arthritis.2 Paradoxical results indicate that Pg has subdued pro-inflammatory effects in immune and non-immune cells (i.e., cytokines/chemokines production) when compared with the effect of other oral bacterial species.3,4 This response, has been linked to several Pg virulence factors such as proteases (gingipains), and lipopolysaccharide (LPS) structural differences compared to other G− species.4,5 The breadth of mechanisms by which Pg could be driving inflammation remains undetermined. Although, recent evidence suggests that Pg, rather than directly being inducing inflammation, it could be indirectly orchestrating changes in the host-oral microbiota interactions (i.e., dysbiosis), which will ultimately drive a chronic inflammatory response leading to further destruction of the periodontal tissues.6 This new model of Pg-driven dysbiosis/periodontitis implies that there could be important variations in both the host response and the oral microbial ecology elicited by Pg that can affect their symbiotic relationships; nonetheless, the mechanisms involved in Pg-induced dysbiosis remain undefined.
Epithelial cells rather than being simply a passive barrier play a critical and active role in maintaining mucosal homeostasis through a constant interaction with the microbiota that lead to the production of an array of antimicrobial and immunoinflammatory factors.7 Therefore, we hypothesized that oral epithelial antimicrobial and immunoinflammatory responses are specifically modulated by oral pathogens such as Pg when compared with oral commensals.
Phospholipase A2 group IIA (PLA2-IIA) is a highly cationic lipolytic enzyme produced by multiple cells (e.g., hepatocytes, platelets, macrophages, and Paneth cells) that belong to a subfamily of ten secreted PLA2 enzymes. PLA2-IIA plays a critical role in infection and inflammation through its potent antimicrobial properties, as well as its role in the pathway for lipid mediators’ production with activation of inflammatory cells such as neutrophils, macrophages, and platelets.8,9,10 Specifically, PLA2-IIA exhibits potent antimicrobial activity due to its high affinity for the phosphatidylethanolamine and phosphatidylglycerol, which are enriched in the membrane of bacteria.11 Consistent with its antimicrobial properties, PLA2-IIA has been recently shown to be an important candidate for host genes that strongly influence the gut microbiota composition,12 as well as a host molecule that can be used by bacterial complexes to regulate species survival in the lungs.13 In addition to its antimicrobial properties, PLA2-IIA induces inflammation by activating neutrophils and macrophages through an M-type receptor (C-type lectin family), enhancing the expression of prostaglandin E2, as well as inducing the production of fatty acids, phospholipids, and release of mitochondrial DNA through its enzymatic activity on extracellular mitochondria expelled from activated platelets.9,10
Herein, using transcriptomic analysis of immunoinflammatory genes in human oral epithelial cells (OECs) we have identified PLA2-IIA as a new molecule through which Pg could be specifically modulating OEC antimicrobial and inflammatory responses. Specifically, we demonstrated that PLA2-IIA expression involves the activation of Notch-1 receptor in OECs by Pg gingipains and the antimicrobial activity of OEC is modulated by Pg through PLA2-IIA. Interestingly, both PLA2-IIA expression and Notch-1 activation showed significant elevations in gingival tissues early during initiation and progression of periodontal disease concurrently with oral dysbiosis, and oral bacterial species showed differential antimicrobial susceptibility to PLA2-IIA. Our findings suggest that a significant increase of PLA2-IIA induced by Pg in OECs may affect the growth and survival of specific bacterial species of the oral microbiome.
Results
PLA2-IIA induction by Pg in OECs
To identify specific OEC responses elicited by Pg that could potentially be associated with disease, transcriptional activity of 520 immunoinflammatory genes with antimicrobial and pro-inflammatory properties were evaluated in OECs exposed to Pg for 24 h using nanostring, and compared with the responses enhanced by the oral commensal S. gordonii (Sg). Interestingly, Pg induced a remarkable increase in the transcription of PLA2-IIA when compared with the response induced by Sg, with expression levels exceeding about 50 times those of classical immunoinflammatory mediators (e.g., cytokines/chemokines and classical antimicrobial peptides such as human beta-defensins) produced by OECs. PLA2G2A was the most up-regulated gene (~3900-fold) elicited by Pg in OECs (Fig. 1). These results were further validated by RT-qPCR where Pg induced about a 100-fold and 500-fold increase in mRNA levels of PLA2G2A at 24 and 48 h after bacterial challenge (Fig. 2a). Consistently with the observed transcriptional response, protein levels of PLA2-IIA were significantly elevated in Pg-exposed OEC lysates (Fig. 2b). Interestingly, strong up-regulation of PLA2-IIA was specifically induced by Pg in OECs. All other oral Gram-positive and Gram-negative bacterial species tested, failed to induce similar responses, while enhancing IL-8 production a standard response to Gram-negative periodontopathogens [e.g., F. nucleatum (Fn), A. actinomycetemcomitans (Aa), and T. forsythia (Tf) (Fig. 2c).
Transcriptional activation of PLA2G2A is strongly induced by P. gingivalis in oral epithelial cells. Immunoinflammatory transcriptome analysis (520 genes) of oral epithelial cells (2.5 × 105 OKF6 cells/well) exposed to the oral pathogen P. gingivalis or the oral commensal S. gordonii [MOI-1:100 for 24 h] was obtained using Nanostring. Arrows indicate location of PLA2G2A and values of fold-change in expression compared with un-stimulated cells are shown
Expression of PLA2-IIA is specifically induced by P. gingivalis in oral epithelial cells. a Transcriptional activation of PLA2G2A was evaluated by RT-qPCR in OKF6 cells exposed to different multiplicities of infection (MOIs) of P. gingivalis (Pg) for 24 and 48 h. b Protein levels of PLA2-IIA induced by P. gingivalis at different MOIs for 24 and 48 h were determined in OKF6 cell lysates by ELISA as described in methods. Normalized amounts of PLA2-IIA in cell lysates per 100 µg of total protein are shown. c Protein levels of PLA2-IIA in cell lysates and IL-8 in supernatants induced by different oral bacterial species at MOI = 1:50 for 48 h in OKF6 cells were determined by ELISA as described in methods section. Normalized amounts of PLA2-IIA in cell lysates per 100 µg of total protein are shown. The mean ± SD of six replicates from each treatment group from a representative experiment of at least two independent experiments are shown. ND Non-detectable. *p ≤ 0.01 when bacteria treated groups where compared with unstimulated (Mock) cells at 24 or 48 h. A.a=Aggregatibacter actinomycetemcomitans
Notch-1 activation is involved in Pg-induced PLA2-IIA expression
The unique ability of Pg for inducing PLA2-IIA expression in OECs, could not be simply explained, by the engagement of pattern recognition receptors such as Toll-like receptors 2 and 4 (TLR2 or TLR4), since most oral bacteria can engage these receptors.14,15 Evidence indicates that Notch-1 signaling is associated with PLA2-IIA production in intestinal epithelial cells in vivo, which appears to play a critical role in gut homeostasis16 and epithelial Notch-1 is a critical regulator of mucosal immune responses.17,18 In addition, it has been demonstrated that Pg stimulates Notch-1 expression in aortic smooth muscle cells (AoSMCs) and osteoblast progenitor cells (OBPCs).19,20 Based on this evidence, we hypothesized that Notch-1 activation in OECs could be a possible target for Pg upregulating PLA2-IIA expression. To explore this, the surface and intracellular expression levels of Notch-1 in OECs cells were first evaluated by flow cytometry. Both, OKF6 and TIGK cells were positive for surface and intracellular Notch-1 expression, with OKF6 cells exhibiting higher mean fluorescence intensity levels than TIGK cells (Supplementary Figure. 1).
The ability of Pg, S. gordonii (Sg), and F. nucleatum (Fn) to activate Notch-1 in OECs was further evaluated by determination of mRNA levels for Hairy and enhancer of split 1 (HES-1), a gene specifically transcribed upon Notch-1 activation.21,22 Pg increased the expression of HES-1 by 3-fold to 9-fold in a dose-dependent manner (Fig. 3a). However, Fn and Sg did not affect HES-1 expression although both increased IL-8 mRNA levels (Fig. 3b). Consistently, nuclear levels of Notch-1 intracellular domain (NICD) were significantly higher in OKF6 cells exposed to Pg but not Fn or Sg compared to un-stimulated cells (Fig. 3c–d). Further, to determine whether Notch-1 activation was involved in Pg-induced PLA2-IIA, the Notch-1/gamma-secretase inhibitors DAPT and JLK6 were used. Protein levels of Pg-induced PLA2-IIA were completely abrogated by both inhibitors at nanomolar (DAPT) and micromolar (JLK6) concentrations without changes in cell viability (Fig. 4a and Supplementary Figure 2). Consistent with these findings, OKF6 cells expressing lower levels of Notch-1 after transfection with a specific siRNA for Notch-1 (Fig. 4b,c), exhibited a significant reduction in the Pg-induced PLA2-IIA mRNA (25-fold less) (Fig. 4d). These findings were consistent with a reduction in Pg-induced PLA2-IIA protein levels, which were almost completely abrogated when compared with cells transfected with the siRNA negative control (Fig. 4d). In contrast, Notch-1 silencing enhanced both constitutive and Pg-induced IL-8 expression (Supplementary Figure 3).
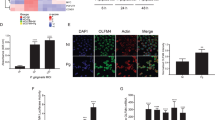
Notch-1 receptor is activated by P. gingivalis in oral epithelial cells. Notch-1 activation was evaluated by quantifying mRNA levels of Hes-1 by RT-qPCR in OKF6 cells exposed to a P. gingivalis or b F. nucleatum (Fn) and S. gordonii (Sg) for 48 h. IL-8 expression was used as a control. c Nuclear levels of Notch-1 intracellular domain (NICD) in OKF6 cells exposed or not to bacteria, were evaluated and quantified by immunofluorescence as described in methods section. Micrographs are showing mock-treated (top) and bacteria-treated OKF6 cells (bottom) for bright field, DAPI, NICD, and DAPI/NICD merge. Higher magnification (20×) of DAPI-NICD merged images is also shown. Scale = 50μm. d Percentage of cells positive for nuclear NICD from panel (c) was determined as described in methods section. The mean ± SD of triplicates from each treatment group from a representative experiment of at least two independent experiments are shown.*p ≤ 0.01 when bacteria-challenged cells were compared with un-stimulated (Mock) cells
Notch-1 activation in oral epithelial cells is involved in P. gingivalis-induced PLA2-IIA. a Effect of specific Notch-1/gamma-secretase inhibitors DAPT and JLK6 on regulating the expression of PLA2-IIA induced by P. gingivalis in oral epithelial cells (OKF6). Pg-challenged cells incubated with the solvent for Notch-1 inhibitors (DMSO) were used as control. Normalized amounts of PLA2-IIA in cell lysates per 100 µg of total protein are shown. b Representative histograms depicting cell surface and intracellular Notch-1 expression levels determined by flow cytometry in OKF6 cells transfected with 20 nM of an siRNA specific for Notch-1 (red line) or its correspondent negative siRNA control (blue line). The gray histogram represents OKF6 cells transfected with a negative siRNA and incubated with an IgG1 isotype control. c Fold-change of cell surface and intracellular Notch-1 expression levels (MFI) determined by flow cytometry and normalized to the MFI from cells incubated with an isotype IgG1 control are shown. d Levels of PLA2-IIA (mRNA and protein) induced by P. gingivalis [1:100] for 48 h in OKF6 cells transfected with siRNA-Notch-1 or the siRNA-negative control. The mean ± SD of triplicates from each treatment group from a representative experiment reproduced at least twice are shown. ND non-detectable. *p ≤ 0.01 DAPT/JLK6 treated cells vs. DMSO-treated cells, and when P. gingivalis challenge was done in cells transfected with Notch-1 siRNA vs. cells transfected with a negative siRNA control
Although Pg can be sensed by TLR2 and TLR4,23 expression and neutralization experiments suggested no effect of these TLRs in Pg-induced Notch-1 activation or PLA2-IIA production by OECs (Supplementary Figure. 4).
Pg gingipains are involved in Notch-1 activation and PLA2-IIA production by OECs
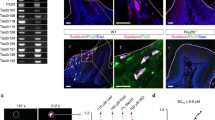
Among several virulence factors, Pg has the unique property to produce three cysteine proteases, Arginine- (rgpA and rgpB) and Lysine- (kgp) gingipains, which degrade many inflammatory mediators such as cytokines/chemokines.24 Gingipains also affect the production of epithelial innate factors (e.g., beta-defensins).25 Since Notch-1 activation involves sequential enzymatic cleavages including the release of the extracellular domain from the cell membrane by the enzyme TNFα-converting enzyme (TACE), followed by a further enzymatic cleavage of the Notch-1 intracellular domain by the gamma-secretase complex, we hypothesized that Pg gingipains could be involved in PLA2-IIA production by OECs through Notch-1 activation. Accordingly, Notch-1 activation was observed only in OECs exposed to Pg wild type (WT) but not in cells challenged with the Pg mutant for all gingipains (Fig. 5a). Nevertheless, both Pg WT and mutant strains increased IL-8 transcription, which was used as a positive control for cell activation. Consistent with these results, significant increase in PLA2-IIA protein levels was observed in OECs exposed to Pg WT but not the Pg mutant (Fig. 5b). Importantly, increased protein levels of IL-8 in OEC supernatants from cells exposed to the Pg triple mutant confirmed its biological activity (Fig. 5c), and as expected, IL-8 levels in supernatants from OECs exposed to Pg WT were not detectable due to degradation associated with Pg gingipains.
Gingipains produced by P. gingivalis are involved in Notch-1 activation and PLA2-IIA production in oral epithelial cells. a Notch-1 activation was determined in OKF6 and TIGK cells by measuring Hes-1 mRNA levels induced by P. gingivalis wild type (WT) or the P. gingivalis mutant strain for all gingipains (rgpA-B-kgp-) after 48 h of stimulation by RT-qPCR. Fold changes were calculated with respect to gene expression levels in cells that were not challenged with bacteria. b PLA2-IIA protein levels produced by OKF6 and TIGK cells in response to P. gingivalis-WT and P. gingivalis-mutant for gingipains [MOI = 1:50] for 48 h were determined by ELISA in cell lysates. Normalized amounts of PLA2-IIA in cell lysates per 100 µg of total protein are shown. c Determination of IL-8 levels by ELISA in cell supernatants (SNs) from either OKF6 or TIGK cells was performed as a control for the biological activity of the P. gingivalis mutant strain. The mean ± SD of triplicates from each treatment group from a representative experiment are shown. *p ≤ 0.05 when bacteria treated groups where compared with unstimulated (Mock) cells. ND non-detectable
Release of Pg-induced PLA2-IIA into the supernatants is enhanced by F. nucleatum
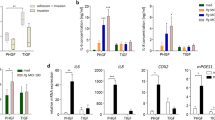
Since oral bacterial species have shown differential ability to induce production and release of pro-inflammatory mediators, we hypothesized that other oral bacterial species could induce release of Pg-induced PLA2-IIA. Detectable levels of PLA2-IIA were found in supernatants from Pg-primed OKF6 cells exposed to Fn but not Sg. Elevated PLA2-IIA levels were observed only in cell lysates from Pg-exposed OECs (Fig. 6).
Release of P. gingivalis-induced PLA2-IIA into the supernatants is enhanced by F. nucleatum. a PLA2-IIA levels were determined by ELISA in cell lysates normalized to total protein (100 μg) and in supernatants from OKF6 cells exposed to other oral bacteria (Fn-F. nucleatum, Sg-S. gordonii at MOI = 1:50) for 24 h, after priming cells with P. gingivalis [1:50 for 24 h]. b Determination of IL-8 levels by ELISA in supernatants was performed as control for biological activity of Fn and Sg. The mean ± SD from two independent experiments by duplicate/triplicate are shown. Interrupted line denotes minimal level of PLA2-IIA detection by ELISA (17 pg/ml). *p ≤ 0.01 when bacteria treated groups where compared with unstimulated (Media) cells
P. gingivalis modulates antimicrobial OEC responses through PLA2-IIA
Since the antimicrobial properties of PLA2-IIA have been broadly described11 we sought to determine if the ability for bacterial killing of OECs could be modulated by Pg through upregulation of PLA2-IIA using L. monocytogenes (Lm) as a susceptible PLA2-IIA target. Pg-challenged OEC extracts significantly reduced the number of Lm colonies forming units (CFUs), whereas unchallenged OECs extracts did not affect bacterial growth. Pre-incubation of Pg-challenged OEC extracts with an anti-PLA2-IIA antibody decreased their ability to reduce the number of CFUs when compared with the effect of its corresponding isotype control (Fig. 7). These Pg-induced OEC antimicrobial responses were similarly induced when rPLA2-IIA was used as a control.
P. gingivalis modulates oral epithelial antimicrobial cell responses through PLA2-IIA. OKF6 cells (1 × 106) were exposed or not to P. gingivalis [1:50] for 48 h and cell lysates obtained with 0.1% Triton in water, in presence of protease inhibitors (PI). Further 40 μl of either OKF6 cell lysates [1 μg/μl total protein] or 10 μg/ml of rPLA2-IIA resuspended in CaCl2 buffer were pre-incubated with either 30 μg/ml of anti-PLA2-IIA antibody or its corresponding isotype control (I. Ctrl.) for 30 min on ice before testing their anti-microbial activity against L. monocytogenes (Lm). Lm (106 in 90 μl broth) was incubated with 10 μl of OKF6 cell lysates or rPLA2-IIA after different treatments for 2 h. Lm incubated only with media, CaCl2 buffer (vehicle for rPLA2-IIA) and Triton + PI (OKF6 cell lysis buffer) were used as controls. Further 25 μl of Lm (1:1000 dilution) were seeded on blood agar plates and colony forming units (CFUs) quantified after 24 h. The mean ± SD of triplicates from each treatment group from a representative experiment of two independent experiments are shown. *p ≤ 0.01, ** p ≤ 0.05 denotes significant reduction in CFUs/ml when different treatments were compared with bacterial growth only with media
PLA2G2A expression, Notch-1 activation and oral microbiome changes during periodontal disease in nonhuman primates (NHPs)
To determine whether Notch-1 activation and PLA2-IIA expression are upregulated in vivo during the pathogenesis of periodontal disease, the ligature induced-periodontitis model in nonhuman primates was used (Fig. 8a). Levels of PLA2G2A were elevated during initiation and progression of disease; however, significant increases in particular at 2 weeks (100-fold) and 1 month (8-fold) after inducing disease with respect to baseline levels were seen (Fig. 8b). Interestingly, a similar pattern of Notch-1 activation determined by expression levels of Hairy/enhancer-of-split related YRPW motif protein 1 (HEY-1, gene specifically transcribed upon Notch-1 activation)26 was observed at 2 weeks (2.5-fold) and 1 month (1.9-fold) (Fig. 8c). The expression levels of both genes were comparable with baseline levels during the clinical resolution phase of the disease. Noteworthy, variations in the abundance of the main oral microbiome phyla were observed during the process of disease, such as reduction in actinobacteria, fusobacteria, and spirochaetes, and increase in the frequency of bacteroidetes and firmicutes as examples (Fig. 8d). Pg abundance increased with disease at 2 weeks (Fig. 8e). Selected oral bacterial species from some Phyla showing reduced numbers during disease (e.g., Actinobacteria: Actinomyces naeslundii, and Fusobacteria: F. nucleatum), also exhibited antimicrobial susceptibility to recombinant human PLA2-IIA (rPLA2-IIA). However, oral bacteria showing antimicrobial resistance belong to phyla showing no changes/increase in abundance (e.g., Firmicutes: Sreptococcus gordonii and Veillonella parvula, and Bacteroidetes: Capnocytophaga sputigena). Pg was resistant to the anti-microbial activity of rPLA2-IIA (Fig. 8f).
PLA2G2A expression, Notch-1 activation and oral microbiome changes during initiation, progression and resolution of periodontal disease in nonhuman primates (NHPs). a The ligature-induced-periodontitis model was used in nonhuman primates M. mulatta (n = 9) as described in methods to determine variations in gingival gene expression by RT-qPCR during initiation (2 weeks and 1 month), progression (3 months) and resolution (2 months after removing ligatures) of periodontal disease. Fold changes in gene expression with respect to baseline levels are shown for b PLA2G2A and c HEY-1 (a marker for Notch-1 activation). Gingival biopsies (n = 9) were pooled in three groups of three samples/each for qRT-PCR analysis. The mean of gene expression is depicted by horizontal bars in each time point. d Variation in abundance of the main oral bacterial Phyla and e P. gingivalis, was determined by 16S rRNA sequencing in oral subgingival plaque samples from NHPs. DNA from plaque samples was isolated (Magnapure LC DNA isolation kit III, Roche), barcoded and amplified using 16S universal primers for V4 region (254 bp amplicon). Amplicons for each sample were purified and pooled in equimolar concentrations and sequenced using Miseq (Illumina). Sequences were clustered into phylotypes based on their sequence similarity and these binned phylotypes were assigned to their respective taxonomic classification using the Human Oral Microbiome Database (HOMD V13). f Antimicrobial effect of recombinant human PLA2-IIA (1 μg/ml for 2 h) in selected oral bacterial species (106) was evaluated by determination of colony forming units (CFUs). Bacteria exposed to 0.1 mg/ml penicillin/streptomycin (Pen/Strep) was used as a positive control and L. monocytogenes (susceptible target for PLA2-IIA) was used as control for the anti-microbial activity of PLA2-IIA in each experiment. The mean ± SD of triplicates from each treatment group from 2–3 independent experiments are shown *p ≤ 0.01, **p ≤ 0.05 when gene expression was compared with baseline levels, and when number of CFUs/ml of Pen/Strep or rPLA2-IIA-treated groups compared vs. Buffer
Discussion
Pg is an oral Gram-negative anaerobe bacterium with the ability to induce dysbiosis and periodontal disease.6 Nevertheless, the mechanisms by which mucosal responses to the oral microbiota in the presence of specific oral pathogenic bacterial species such as Pg could abrogate the host-microbe symbiotic relationship leading to dysbiosis and later to periodontitis remain not fully understood.
Possible mechanisms associated with the onset of dysbiosis could be a direct impact of certain bacterial species competing or killing other members of the bacterial communities through the production of antimicrobial molecules such as bacteriocins,27 or the modulation of host mucosal antimicrobial and inflammatory responses that can impact the growth or persistence of susceptible species within these bacterial communities.28 Indeed, previous studies indicated that subversion of neutrophil responses by Pg that involve C5aR and TLR2 receptor activation and the serine protease (Ser B) that decreased IL-8 expression by OECs can play a role in oral dysbiosis.29 Nevertheless, whether or not Pg could specifically modulate antimicrobial and immunoinflammatory responses in OECs with potential to impact the abundance of certain oral bacterial species remains unknown.
A transcriptome analysis of antimicrobial and inflammatory genes in OECs indicated that Pg is a strong inducer of PLA2-IIA expression and this response seems to be specific, since other bacterial species failed to induce similar responses. Production of PLA2-IIA is induced by other bacterial species such as Neisseria meningitides in macrophages and Pseudomonas aeruginosa in human bronchial epithelial cells through mechanisms that involve the pili and a type III secretion system respectively.13,30 Nevertheless, the cellular and molecular mechanisms by which PLA2-IIA expression is modulated in mammalian cells by bacterial components remain unclear. Herein, we demonstrated that Pg is able to induce the production of PLA2-IIA in a mechanism that involves Notch-1 receptor activation in OECs. To our knowledge, Pg could be one of the few microorganisms reported to date, including Hepatitis B and C viruses and Kaposi’s sarcoma herpesvirus31,32 that have the ability to activate the Notch-1 receptor. Notch-1 activation regulates different cell responses such as proliferation, differentiation and apoptosis as well as seems to be a critical regulator of the mucosal immune responses.17 As a consequence, activation of Notch-1 specifically by Pg could be contributing in the pathogenesis of diseases in which this oral pathogen has been previously linked, such as periodontal disease, atherosclerosis, and oral cancer.
Although the precise molecular mechanisms by which Pg is activating Notch-1 remain to be elucidated, we showed that gingipains produced by Pg seem to be involved. In particular, the sequence of the Notch-1 extracellular domain (NECD) that is in close proximity with the cell membrane and interacts non-covalently with the NICD has a sequence of four Arginine amino acids that could be targeted by Pg rgp-A and rgp-B gingipains.33 Thus, it is likely that gingipains and most specifically Arginine-gingipains could be inducing a non-canonical Notch-1 activation, enhancing an initial cleavage of the NECD, with the consequent activation of the gamma-secretase complex and further release and nuclear translocation of the NICD, whereby it can start transcription of specific genes including PLA2-IIA. Expression of HES-1 and HEY-1 have been shown to be regulated through Notch-1-independent pathways such as c-jun N-terminal kinase (JNK) signaling, which is also activated by Pg in OECs.34,35 Nevertheless, the use of additional approaches (e.g., nuclear levels of NICD or siRNA Notch-1 silencing) reinforce the observation of a Pg-induced Notch-1 activation using these markers.
Previous studies have shown that TLR2 activation by Pg induces the expression of other inflammatory and antimicrobial mediators by OECs.23 The lack of a role for TLR-2 in Pg-induced PLA2-IIA is consistent with the specificity of these oral epithelial responses induced by Pg, while other oral commensal and pathogenic bacteria, which also activate TLR-2, failed to induce similar PLA2-IIA responses. OECs have been shown to respond to the TLR4 ligand (i.e., purified E. coli LPS), and blocking TLR4 reduced the expression of the CCR5 receptor in OECs.36 In this study, flow cytometry analysis showed that neither OKF6 nor TIGKs constitutively express TLR4. These findings are consistent with low/undetectable TLR4 mRNA expression levels in primary OECs compared with hematopoietic cells such as macrophages.37 Although the potential role of TLR4 in Pg-induced PLA2-IIA could not be completely ruled out at this point (given that TLR4 expression may be induced after bacterial challenge), the contribution of this pathway remains questionable, given that other oral Gram-negative bacteria such as F. nucleatum, T. forsythia and A. actinomycetemcomitans also failed to induce PLA2-IIA in OECs.
The anti-microbial properties of PLA2-IIA have been described both in vitro and in vivo.11,12,13 Herein, as a proof of concept, we demonstrated in vitro that Pg enhanced the antimicrobial properties of OECs stimulating the production of PLA2-IIA, and oral bacterial species have differential anti-microbial susceptibility to rPLA2-IIA. These findings suggest that modulation of PLA2-IIA levels in OECs by Pg could be contributing to modify the abundance of specific bacterial species within the oral microbiome. Nevertheless, this needs to be mechanistically demonstrated in future in vivo studies.
Even though PLA2-IIA is an important component in gastrointestinal and respiratory mucosal inflammatory disorders,10,38 there are surprisingly few reports demonstrating any variations in the expression levels of PLA2-IIA in oral mucosa or gingival crevicular fluid during health or periodontitis.39,40 Using the ligature-induced-periodontitis model in nonhuman primates we found that significant increases in the gingival expression of PLA2-IIA occur early during the initiation phase of the disease. Accordingly, a similar expression pattern was observed for Notch-1 activation with significant elevations occurring very rapidly after disease induction. These findings suggest that up-regulation of the Notch-1/PLA2-IIA axis in gingival tissues could be involved in early events of the pathogenesis of periodontitis. These observations are consistent with previous studies showing elevated levels of PLA2-IIA associated with several chronic inflammatory disorders such as rheumatoid arthritis, inflammatory bowel disease, atherosclerosis, as well as diet-induced metabolic syndrome.41,42
While the role of PLA2-IIA in the pathogenesis of periodontal disease in vivo remains to be determined, previous evidence supports the hypothesis that this enzyme may play a significant role. For example, mouse strains (e.g., C57BL/6, 129/J) that have a natural mutation in PLA2G2A gene (leading to the production of an inactive enzyme) are more resistant to Pg-induced periodontitis, than mouse strains (e.g., BALB/c, DBA/2J) that express a functional PLA2-IIA enzyme.43,44 Therefore, a non-functional PLA2-IIA in resistant mice strains may be preventing potential Pg-induced responses mediated by PLA2-IIA that could lead to dysbiosis and further periodontitis. Future mechanistic studies using different mice strains and PLA2-IIA transgenic mice are warranted to test and validate in vivo the potential role of this enzyme in oral dysbiosis and periodontal disease.
Overall, we have demonstrated that the oral pathogen Pg is a strong activator of PLA2-IIA in OECs through a mechanism that involves Notch-1 activation by gingipains, and activation of PLA2-IIA increases the antimicrobial properties of OECs. Moreover, the Notch-1/PLA2-IIA axis is upregulated early in the periodontal disease process, and oral bacterial species exhibit differential antimicrobial susceptibility to PLA2-IIA, which seems to correlate with disease-associated oral microbiome changes. These findings support previous studies reporting a positive correlation between Notch-1 activation and PLA2-IIA expression in intestinal epithelial cells16, and provide new mechanistic information about the regulation of epithelial antimicrobial and pro-inflammatory responses modulated by oral pathogenic bacteria through the Notch-1/PLA2-IIA axis that could be possibly contributing to disease.
PLA2-IIA with its targeted antimicrobial activity and ability to modulate immunoinflammatory responses, combined with P. gingivalis commandeering the Notch-1 receptor pathway, makes this an interesting and novel potential strategy used by this oral pathogen to modify the local ecology that triggers early changes in the bacteria-host homeostatic interactions leading to disease.
Methods
Human cell types and bacteria
The immortalized keratinocyte cell line OKF6, established by ectopic expression of the telomerase catalytic subunit (hTERT) in cells from normal oral mucosal epithelium45 and the telomerase immortalized human gingival epithelial cells (TIGK)46 were used in these studies. Both OEC lines were cultured in antibiotic-free, serum-free keratinocyte medium (Ker-SFM, Gibco, Carlsbad, CA) supplemented with 0.2 ng/ml human recombinant epidermal growth factor and 25 μg/ml bovine pituitary extract. THP-1 cells were grown in RPMI 1640 (Sigma) with 2 mM L-glutamine and 10% FBS (Seradigm, Radnor, PA). All cells were grown in 5% CO2 in air at 37 °C.
The following bacterial strains were used in these studies: Porphyromonas gingivalis 381 (Pg), Sreptococcus gordonii ATCC10558 (Sg), Streptococcus sanguinis ATCC10556 (Ss), Actinomyces naeslundiiATCC49340 (An), Aggregatibacter actinomycetemcomitansJP2 (Aa), Tannerella forsythia ATCC43037 (Tf), Fusobacterium nucleatum ATCC25586 (Fn), Capnocytophaga sputigena ATCC33612, Veillonella parvula ATCC10790, and Listeria monocytogenes EDGe strain. All bacterial strains, were initially grown on blood agar plates (BBL, Becton Dickinson, Sparks, MD, USA) from a frozen stock and incubated in appropriate aerobic or anaerobic conditions during 24 h or 3 days, respectively. Further, a liquid culture was started for each strain in 3 ml of Brain Heart Infusion Broth alone or supplemented with 5 μg/ml Hemin and 1 μg/ml Menadione in the case of Pg. Cells were incubated for 24 h and then overnight stationary cultures were inoculated in fresh BHI broth and allowed to reach logarithmic phase for 3–4 h. Finally, bacteria were counted in a hemocytometer by microscopy, and a cell suspension adjusted to 1 × 107 cells/ml in Ker-SFM with supplements for further experiments challenging OECs. T. forsythia was grown in 2.5 ml of ATTC medium 1921 supplemented with 10 µg/ml of N-acetyl muramic acid (NAM). The Pg mutant for gingipains (rgpA-rgpB-kgp) created under the Pg381 background was grown in blood agar plates and Trypticase Soy broth supplemented with Yeast extract, Hemin 5 μg/ml and Vitamin K 1 μg/ml in presence of Erythromycin 10 μg/ml and Tetracycline 2 μg/ml.
Immunoinflammatory transcriptome analysis by nanostring
Immunoinflammatory transcriptome analysis of OECs exposed to the oral pathogen Pg or the oral commensal S. gordonii [MOI-1:100 for 24 h] was performed using a set of 520 genes defined by the nCounter Human Immunology Kit panel (Nanostring, Seattle, WA). Briefly, after bacterial exposure, total mRNA was isolated using the PureLink RNA Mini kit (Life Technologies, NY, USA). Further, 100 ng of RNA were hybridized with the reporter code set beads for a final volume of 30 μl at 65 °C for 12 h. Finally, the samples were processed on the Nanostring Cell Prep Station and data collected using the Nanostring Digital Analyzer version (Nanostring Technologies, Seattle, WA, USA).
Quantitative RT-PCR for human PLA2-IIA, HES-1, IL-8
Transcriptional activation of PLA2G2A, HES-1, and IL-8 was evaluated by RT-qPCR in OKF6 cells after different treatments with oral bacteria. RNA was extracted using PureLink™ RNA Mini Kit (Invitrogen) and measured on a NanoDrop (Thermo, Waltham, MA) for concentration and quality. Genomic DNA was removed using PerfeCTa® DNase I (Quanta, Beverly, MA). RNA was then reverse-transcribed using qScript™ cDNA Synthesis Kit (Quanta). qPCR assays were run on 96 well plates using PerfeCTa® SYBR® Green SuperMix (Quanta) and read on LightCycler480 (Roche, Basel, Switzerland) using the following cycling conditions: Initial denaturation: 95 °C, 3 min. PCR cycling 45 cycles (95 °C, 15 s, 60 °C, 30 s). The sequences for all used primers are shown in Table 1. Relative quantification method (based on 2−∆∆CT) was used to calculate fold differences using GAPDH as an endogenous control.
Enzyme linked immunosorbent assays
Protein levels of PLA2-IIA and IL-8 either in cell lysates or supernatants were determined by ELISA using the PLA2 (human type IIA EIA) immunometric assay kit (Cayman Chemicals Company, Ann Arbor, MI, USA) and an IL-8 ELISA MAX standard kit (BioLegend, San Diego, CA). For cell lysates, after different treatments cell monolayers were washed with 1 × PBS and lysed with 300 μl RIPA buffer for 30 min on ice followed by centrifugation at 14,000 rpm at 4 °C. Supernatants were stored at −20 °C for ELISA analysis. Determination of total protein was performed using the DC protein assay kit (BIO-RAD, Hercules, CA, USA) and PLA2-IIA amounts obtained by ELISA were normalized to a constant amount of protein for all samples.
Flow cytometry analysis and immunofluorescence
For analysis of Notch-1 expression, confluent cell cultures of OKF6 and TIGK cells grown in 75 cm2 flasks were dissociated using Trypsin-EDTA solution for 5 min, and washed at 1200 rpm for 5 min. Further, 1 × 105 cells in 100 μL FACS buffer were then either incubated with 5 μL monoclonal IgG antibody against Notch-1 conjugated with phycoerythrin (anti-Notch-1-PE) (recognizes the Notch-1 extracellular domain) or an IgG isotype control (R&D Systems, Minneapolis, MN) undiluted for 30 min on ice. Cells were then washed twice with PBS (1200 rpm, 5 min), and resuspended in 100 μL FACS buffer for further analysis. For intracellular Notch-1 staining, a similar number of cells were fixed for 15 min with 2% paraformaldehyde solution at room temperature and then washed and permeabilized using 1× intracellular staining permeabilization wash buffer (BioLegend, San Diego, CA) before adding the corresponding antibodies.
OKF6 (1 × 105) cells grown on coverslips and exposed to Pg [1:100] for 24 h were fixed, permeabilized and stained with anti-cleaved Notch-1 (NICD) (ab8925; abcam, Cambridge, UK) dilution 1:200 for 1 h, followed by goat anti-rabbit AF467 (Invitrogen, Carlsbad, CA) at 1:500 dilution for 1 h. DAPI was used to stain the nuclei. Images were captured, and percentage of NICD positive nuclei from three random regions of interest per slide analyzed using the Nikon TiS fluorescent microscope, and NIS-Elements Basic Research software (Tokyo, Japan).
Notch-1 inhibitors and siRNA silencing
OKF6 cells (2 × 105 cells/well) were seeded in 24-well plates in 1 ml of media and pre-incubated with the gamma-secretase/Notch-1 inhibitors DAPT (EMD Millipore, Billerica, MA) or JLK6 (Tocris Bioscience, Bristol, UK) diluted in DMSO for 1 h before challenge with Pg [1:50] for 48 h. Supernatants were harvested for further analysis and cells lysed for PLA2-IIA analysis by ELISA.
For transfection experiments, OKF6 cells (1 × 105) were seeded on 12 well plates in 1 mL Ker-SFM overnight. Then, cells were transfected with either Notch 1 small interfering RNA (siRNA), or a negative control siRNA (Invitrogen, Carlsbad, CA). Briefly, 2 μL per well Lipofectamine RNAiMAX Reagent (Invitrogen) or 2 μl of each siRNA were independently diluted in Opti-MEM medium. Each diluted siRNA was added to diluted Lipofectamine RNAiMAX reagent (1:1 ratio) and incubated for 5 min at room temperature. The siRNA-lipid complexes were then added dropwise to the cells (final concentration of siRNA = 20 nM) and cells were incubated for 24 h at 37 °C before being harvested for Notch-1 expression analysis.
Antimicrobial effect of rPLA2-IIA and Pg-induced PLA2-IIA
Oral bacterial species (106) growing exponentially in BHI broth were individually exposed to human recombinant PLA2-IIA (1 μg/ml) (R&D Systems, Minneapolis, MN, USA) or its vehicle (CaCl2 buffer) for 2 h at 37 °C in appropriate aerobic or anaerobic conditions and further seeded and incubated in blood agar plates to determine colony forming units (CFUs). Bacteria exposed to 0.1 mg/ml penicillin/streptomycin was used as a positive control and L. monocytogenes (susceptible target for PLA2-IIA) used as control for anti-microbial activity of PLA2-IIA.
To test the antimicrobial effect of Pg-induced PLA2-IIA, OKF6 cells (106) were exposed or not to Pg [1:50] for 48 h and cell lysates obtained with 0.1% Triton in water, in presence of protease inhibitors (PI) (Complete Mini-EDTA-free, Roche Diagnostics, Indianapolis, IN, USA). Then, 40 μl of OKF6 cell lysates [1 μg/μl total protein] or 10 μg/ml of rPLA2-IIA resuspended in CaCl2 buffer were pre-incubated with anti-PLA2-IIA antibody or its corresponding isotype control (30 μg/ml) for 30 min on ice. Further Lm (106 in 90 μl of broth) was incubated with 10 μl of either OKF6 cell lysates or rPLA2-IIA after different treatments for 2 h. Lm incubated only with media, CaCl2 buffer (vehicle for rPLA2-IIA) and Triton + PI (OKF6 cell lysis buffer) were used as controls. Finally, 25 μl of Lm (1:1000 dilution) were seeded on blood agar plates and CFUs quantified after 24 h.
Ligature-induced periodontal disease, gingival tissue and plaque sample analysis
Rhesus monkeys (Macaca mulatta) (n = 9, 6 females and 3 males) housed at the Caribbean Primate Research Center (CPRC) at Sabana Seca, Puerto Rico, were used in these studies. The range of age was 12–14 years old (Mean = 12.6 ± 0.9). Following a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Puerto Rico, a single investigator periodontally examined anesthetized animals as we have previously described.47,48 At the initiation of the study, all animals were evaluated clinically for periodontal disease, and only animals with mean bleeding on probing (BOP) of ≤1 and mean pocket depths (PD) < 3 mm were entered into the study.
After taking the baseline gingival samples, ligatures were placed on the second premolar and first and second molars of both maxillary quadrants. Further, clinical evaluation for ligated sites was obtained and a buccal gingival papilla from each animal was taken using a standard gingivectomy technique at each time point as we and others have previously reported.49,48 Samples were maintained frozen at −80 °C in RNAlater solution until real time RT-qPCR analysis.48 Isolated RNA from three different animals was pooled (total of three groups of three pooled samples/time point) and a total of 1 μg was used for each cDNA synthesis reaction. Sequences for the forward and reverse primers for PLA2G2A, HEY-1, and GAPDH are shown in Table 1. The relative quantification analysis was performed with the LightCycler480 software (Roche, IN). The concentration ratios for the target genes were calculated by normalizing to the housekeeping gene GAPDH. Oral subgingival plaque samples were also analyzed by 16S rRNA sequencing using Miseq Illumina system (Illumina, San Diego, CA, USA).
Statistical analysis
For the in vitro experiments, ANOVA was used for comparisons among experimental groups, and unpaired student’s t test for comparisons between two groups. Significant variation of gene expression in gingival tissues from nonhuman primates at different time was evaluated using a one way ANOVA and a two sample t-tests.
Change history
22 February 2019
The sequence for the Reverse primer used to amplify the human gene PLA2G2A presented in table 1 is incorrect. The following, is the correct sequence: Reverse 5’ - GCTCCCTCTGCAGTGTTTATT -3
References
Socransky, S. S. et al. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144 (1998).
Hajishengallis, G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15, 30–44 (2015).
Darveau, R. P., Belton, C. M., Reife, R. A. & Lamont, R. J. Local chemokine paralysis, a novel pathogenic mechanism for porphyromonas gingivalis. Infec. Immun. 66, 1660–1665 (1998).
Darveau, R. P., Arbabi, S., Garcia, I., Bainbridge, B. & Maier, R. V. Porphyromonas gingivalis lipopolysaccharide is both agonist and antagonist for p38 mitogen-activated protein kinase activation. Infec. Immun. 70, 1867–1873 (2002).
Imamura, T. The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol. 74, 111–118 (2003).
Hajishengallis, G. et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10, 497–506 (2011).
Peterson, L. W. & Artis, D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14, 141–153 (2014).
Murakami, M., Sato, H., Miki, Y., Yamamoto, K. & Taketomi, Y. A new era of secreted phospholipase A(2). J. Lipid Res. 56, 1248–1261 (2015).
Duchez, A. C. et al. Platelet microparticles are internalized in neutrophils via the concerted activity of 12-lipoxygenase and secreted phospholipase A2-IIA. Proc. Natl Acad. Sci. USA 112, E3564–E3573 (2015).
Bidgood, M. J., Jamal, O. S., Cunningham, A. M., Brooks, P. M. & Scott, K. F. Type IIA secretory phospholipase A2 up-regulates cyclooxygenase-2 and amplifies cytokine-mediated prostaglandin production in human rheumatoid synoviocytes. J. Immunol. 165, 2790–2797 (2000).
Nevalainen, T. J., Graham, G. G. & Scott, K. F. Antibacterial actions of secreted phospholipases A2. Review. Biochim. Biophys. Acta 1781, 1–9 (2008).
Brodziak, F., Meharg, C., Blaut, M. & Loh, G. Differences in mucosal gene expression in the colon of two inbred mouse strains after colonization with commensal gut bacteria. PLoS ONE 8, e72317 (2013).
Pernet, E. et al. Pseudomonas aeruginosa eradicates Staphylococcus aureus by manipulating the host immunity. Nat. Commun. 5, 5105 (2014).
Peyret-Lacombe, A., Brunel, G., Watts, M., Charveron, M. & Duplan, H. TLR2 sensing of F. nucleatum and S. sanguinis distinctly triggered gingival innate response. Cytokine 46, 201–210 (2009).
Gonzalez, O. A., Li, M., Ebersole, J. L. & Huang, C. B. HIV-1 reactivation induced by the periodontal pathogens Fusobacterium nucleatum and Porphyromonas gingivalis involves Toll-like receptor 2 [corrected] and 9 activation in monocytes/macrophages. Clin. Vaccin. Immunol. 17, 1417–1427 (2010).
Okamoto, R. et al. Requirement of Notch activation during regeneration of the intestinal epithelia. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G23–G35 (2009).
Mathern, D. R. et al. Mouse and human Notch-1 regulate mucosal immune responses. Mucosal Immunol. 7, 995–1005 (2014).
Hu, X. et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity 29, 691–703 (2008).
Zhang, B. et al. The periodontal pathogen Porphyromonas gingivalis changes the gene expression in vascular smooth muscle cells involving the TGFbeta/Notch signalling pathway and increased cell proliferation. BMC Genom 14, 770 (2013).
Xing, Q. et al. Porphyromonas gingivalis lipopolysaccharide inhibits the osteoblastic differentiation of preosteoblasts by activating Notch1 signaling. J. Cell Physiol. 225, 106–114 (2010).
Guruharsha, K. G., Kankel, M. W. & Artavanis-Tsakonas, S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat. Rev. Genet. 13, 654–666 (2012).
Jarriault, S. et al. Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol. Cell. Biol. 18, 7423–7431 (1998).
Darveau, R. P. et al. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect. Immun. 72, 5041–5051 (2004).
Potempa, J., Sroka, A., Imamura, T. & Travis, J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr. Protein Pept. Sci. 4, 397–407 (2003).
Chung, W. O., Hansen, S. R., Rao, D. & Dale, B. A. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J. Immunol. 173, 5165–5170 (2004).
Maier, M. M. & Gessler, M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem. Biophys. Res. Commun. 275, 652–660 (2000).
Sana, T. G. et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl Acad. Sci. USA 113, E5044–E5051 (2016).
Zenewicz, L. A. et al. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J. Immunol. 190, 5306–5312 (2013).
Lamont, R. J. & Hajishengallis, G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 21, 172–183 (2015).
Touqui, L. et al. Neisseria meningitidis pili induce type-IIA phospholipase A2 expression in alveolar macrophages. FEBS Lett. 579, 4923–4927 (2005).
Kongkavitoon, P., Tangkijvanich, P., Hirankarn, N. & Palaga, T. Hepatitis B virus HBx activates Notch signaling via delta-like 4/Notch1 in hepatocellular carcinoma. PLoS ONE 11, e0146696 (2016).
Lan, K., Choudhuri, T., Murakami, M., Kuppers, D. A. & Robertson, E. S. Intracellular activated Notch1 is critical for proliferation of Kaposi’s sarcoma-associated herpesvirus-associated B-lymphoma cell lines in vitro. J. Virol. 80, 6411–6419 (2006).
Ranganathan, P., Weaver, K. L. & Capobianco, A. J. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat. Rev. Cancer 11, 338–351 (2011).
Curry, C. L., Reed, L. L., Nickoloff, B. J., Miele, L. & Foreman, K. E. Notch-independent regulation of Hes-1 expression by c-Jun N-terminal kinase signaling in human endothelial cells. Lab. Invest. 86, 842–852 (2006).
Wang, Q. et al. FOXO responses to Porphyromonas gingivalis in epithelial cells. Cell. Microbiol. 17, 1605–1617 (2015).
Giacaman, R. A. et al. Porphyromonas gingivalis induces CCR5-dependent transfer of infectious HIV-1 from oral keratinocytes to permissive cells. Retrovirology 5, 29 (2008).
Neiva, K. G., Calderon, N. L., Alonso, T. R., Panagakos, F. & Wallet, S. M. Type 1 diabetes-associated TLR responsiveness of oral epithelial cells. J. Dent. Res. 93, 169–174 (2014).
Yamaguchi, O. et al. Correlation between serum phospholipase A(2) IIA levels and histological activity in patients with ulcerative colitis. Int. J. Colorectal Dis. 17, 311–316 (2002).
Ishida, H., Shinohara, H., Nagata, T., Nishikawa, S. & Wakano, Y. Phospholipase A(2) activity in gingival crevicular fluid from patients with periodontal disease: a possible marker of disease activity. Mediat. Inflamm. 3, 17–21 (1994).
Shinohara, H. et al. Group II phospholipase A(2) in human gingiva with periodontal disease. Mediat. Inflamm. 4, 95–97 (1995).
Ibeas, E., Fuentes, L., Martin, R., Hernandez, M. & Nieto, M. L. Secreted phospholipase A2 type IIA as a mediator connecting innate and adaptive immunity: new role in atherosclerosis. Cardiovasc. Res. 81, 54–63 (2009).
Iyer, A. et al. An inhibitor of phospholipase A2 group IIA modulates adipocyte signaling and protects against diet-induced metabolic syndrome in rats. Diabetes 61, 2320–2329 (2012).
Baker, P. J., Dixon, M. & Roopenian, D. C. Genetic control of susceptibility to Porphyromonas gingivalis-induced alveolar bone loss in mice. Infect. Immun. 68, 5864–5868 (2000).
Kennedy, B. P. et al. A natural disruption of the secretory group II phospholipase A2 gene in inbred mouse strains. J. Biol. Chem. 270, 22378–22385 (1995).
Dickson, M. A. et al. Human keratinocytes that express hTERT and also bypass ap16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 20, 1436–1447 (2000).
Moffatt-Jauregui, C. E. et al. Establishment and characterization of a telomerase immortalized human gingival epithelial cell line. J. Periodontal Res. 48, 713–721 (2013).
Ebersole, J. L., Steffen, M. J., Gonzalez-Martinez, J. & Novak, M. J. Effects of age and oral disease on systemic inflammatory and immune parameters in nonhuman primates. Clin. Vaccin. Immunol. 15, 1067–1075 (2008).
Ebersole, J. L. et al. Cytokine gene expression profiles during initiation, progression and resolution of periodontitis. J. Clin. Periodontol. 41, 853–861 (2014).
Moritz, A. J., Cappelli, D., Lantz, M. S., Holt, S. C. & Ebersole, J. L. Immunization with Porphyromonas gingivalis cysteine protease: effects on experimental gingivitis and ligature-induced periodontitis in Macaca fascicularis. J. Periodontol. 69, 686–697 (1998).
Acknowledgements
We would like to thank Drs. Richard Lamont (University of Louisville), Ann Progulske-Fox (University of Florida) and Sarah D’Orazio (UK) for their generosity in sharing the TIGK cells, Pg mutant strains for gingipains, and Lm strain respectively. We also thank the Genetics Core from University of Kentucky for their support with Nanostring and 16S sequencing experiments. NIH/NIDCR Grant DE024804 and NIH/NIGMS Grant P20GM103538 supported this research.
Author information
Authors and Affiliations
Contributions
A.A., Y.A., S.K. designed and performed in vitro experiments, analyzed data, and wrote manuscript; A.K. assisted in vitro experiments; M.J.N., L.O., J.G., M.M., and J.L.E. contributed designing and conducting the nonhuman primate experiments as well as analyzing the data; A.J.S. supported the experimental design and statistical analysis; O.A. design the study, supervised and performed experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Al-Attar, A., Alimova, Y., Kirakodu, S. et al. Activation of Notch-1 in oral epithelial cells by P. gingivalis triggers the expression of the antimicrobial protein PLA2-IIA. Mucosal Immunol 11, 1047–1059 (2018). https://doi.org/10.1038/s41385-018-0014-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-018-0014-7
This article is cited by
-
The oral microbiome in autoimmune diseases: friend or foe?
Journal of Translational Medicine (2023)
-
Transcriptomic phases of periodontitis lesions using the nonhuman primate model
Scientific Reports (2021)
-
Regulation of olfactomedin 4 by Porphyromonas gingivalis in a community context
The ISME Journal (2021)
-
The oral microbiota: dynamic communities and host interactions
Nature Reviews Microbiology (2018)