Abstract
A balanced gut microbiota is important for human health, but the mechanisms that maintain homeostasis are incompletely understood. Recent insights suggest the host plays a key role in shaping its gut microbiota to be beneficial. While host control in the small intestine curbs bacterial numbers to avoid competition for simple sugars and amino acids, the host limits oxygen availability in the large intestine to obtain microbial fermentation products from fiber. Epithelial cells are major players in imposing ecological control mechanisms, which involves the release of antimicrobial peptides by small-intestinal Paneth cells and maintenance of luminal anaerobiosis by epithelial hypoxia in the colon. Harnessing these epithelial control mechanisms for therapeutic means could provide a novel lynchpin for strategies to remediate dysbiosis.
Similar content being viewed by others
Introduction
Since the first demonstration of causality by Koch’s postulates,1 the idea that microorganisms cause diseases has been a leading concept in pathogenesis. Frank pathogens can cause sickness in individuals with an intact immune system, which supports the idea that microbial virulence factors are the principal drivers of the disease process. Although the downstream host response to infection can contribute to the development of symptoms, disease caused by frank pathogens is not due to an underlying host defect. Therefore treating symptoms produced by the host immune response can provide benefit, but curing the disease requires eradicating its cause, the microbial pathogen.
The idea that microorganisms cause diseases is also a popular concept driving research on the contribution of the host-associated microbial community (microbiota) to disease.2,3 The view that an imbalance in the microbial community (dysbiosis) is an underlying cause of sickness provides a rationale for establishing commensal Koch’s postulates for microbiota research4 and raises hope that administering fecal microbiota transplants (FMTs) to eradicate a microbial imbalance could be developed as a cure.5,6 The latter approach is successful in treating antibiotic-associated colitis,7 because here the FMT directly targets the underlying defect, namely an antibiotic-mediated disruption of the gut microbiota.8 However, lasting benefits of FMT remain to be established for diseases associated with dysbiosis developing independently of antibiotic treatment.9,10,11
An alternative view holds that dysbiosis exacerbates disease, but that it is secondary to an underlying defect in the host,8 which impairs control over the microbial ecosystem.12 The defect in the host’s ability to control the microbial ecosystem can lead to downstream consequences, such as changes in the microbial metabolite landscape, which can contribute to the development of symptoms. Thus, remediating dysbiosis with probiotics or FMTs can provide benefit, but curing the disease will require eradicating its cause by restoring host control over the microbial ecosystem.12,13
This review reports our current understanding of how the host maintains control over its microbial ecosystem. The main focus will be on the human gastrointestinal tract, because this site harbors the largest microbial community, whose disturbance is associated with many human illnesses, including inflammatory bowel diseases,14 colorectal cancer,15,16 athrosclerosis,17 and even neurological disorders.18,19
Host control at its extreme
Ecological theory suggests that hosts are under strong natural selection to shape their microbiota to be beneficial.13 Host control over the microbial community structure is particularly striking for the light organ of the bobtail squid, which is permissive for only a single-bacterial species, the luminous Vibrio fischeri (phylum Proteobacteria) (Fig. 1a).20 The light organ acquires V. fischeri horizontally,21 which suggests that the bobtail squid must select the correct bacterial species from some 1800 bacterial species present in seawater.22 The underlying species–specific selection mechanisms are not fully resolved, but include a number of antimicrobial factors produced by the host.23,24,25 Interestingly, luciferase, the enzyme catalyzing light production, is required for colonization of the light organ,26 presumably because luminescence consumes oxygen, thereby protecting V. fischeri from bactericidal phenoloxidase activity, which is produced by the host to control the environemnt.27 Through these mechanisms, the host limits access to its light organ to a single-bacterial species producing luminescence, which in turn provides benefit by supporting the bobtail squid’s camouflaging behavior termed counterillumination.
Host control can steer the microbial community composition towards dominance of a single genus. a Antimicrobial factors produced by the bobtail squid limit colonization of its light organ to a single species, Vibrio fischeri.20 b Host control mechanisms direct the composition of the cervical and vaginal microbiota in the majority of reproductive-aged women toward a dominance of Lactobacillus species.28,29 c Maternal control imposed through milk oligosaccharides drives a dominance of Bifidobacterium species in the infant gut microbiota.34
Although, our body does not constrain any of its microbial communities to just a single-bacterial species, the female reproductive tract brings our ability to shape the microbiota composition into plain sight. In the majority of reproductive-aged women, >97.5% of the bacteria in the cervical and vaginal microbiota belong to one or more species of just a single genus, Lactobacillus (phylum Firmicutes) (Fig. 1b).28,29 This dominance of a single genus illustrates that similar to the bobtail squid, the human host can keep its microbiota on a very short leash. The proposed mechanism selecting for a Lactobacillus-dominated microbiota is hormonal control by estrogen,30 which coordinates α-amylase-mediated catabolism of host-derived glycogen into maltose and maltotriose. In turn these host-derived oligosaccharides promote fermentative growth by Lactobacillus species.31 This host-mediated nutritional selection for a Lactobacillus-dominated vaginal microbiota is thought to provide benefit by imparting a barrier to infection with vaginal pathogens.32
Host control over microbes also becomes obvious during nursing, a maternal influence that profoundly shapes the composition of the colonic microbiota during infancy. Human milk oligosaccharides are complex carbohydrates that cannot be broken down by enzymes in the upper gastrointestinal tract of the infant and reach the colon,33 where they foster development of a microbiota dominated by species belonging to the genus Bifidobacterium (phylum Actinobacteria), which account for up to 70% of the bacteria present in feces (Fig. 1c).34 Consistent with the idea that human milk oligosaccharides selectively feed members of the genus Bifidobacterium, the infant-associated B. Iongum subspecies infantis contains a carbohydrate utilization gene cluster for fermentative breakdown of these nutrients,35 which is absent from the genome of species that do not dominate the microbiota of breast-fed infants.36 By shaping the composition of the gut microbiota during a critical period when the immune system is still developing, maternal control mediated through human milk oligosaccharides provides benefit for infant health.37
While the examples above illustrate that the host can profoundly shape the microbiota composition by imposing domination of a single genus (Fig. 1), host control over the microbiota composition is not apparent from fecal microbiota analysis of human adults, which provides no evidence for any bacterial species being under positive selection.38 Once nutritional control mediated through milk oligosaccharides is relieved at weaning, the dominance of the genus Bifidobacterium gives way to a diverse microbial community, predominantly composed of obligate anaerobic bacteria belonging to the phyla Firmicutes and Bacteroidetes. On the species level, the human gut microbiota composition shifts strongly with changes in the diet,39 thus supporting the idea that diet is one of the main drivers in shaping the gut microbiota.40 In the absence of any blatant signs of host control over the microbiota in adults, mechanisms that maintain a balanced colonic microbial community during gut homeostasis have remained elusive. In the following sections we will review recent evidence suggesting that host control over the microbial ecosystem is also a prominent feature in the adult colon, but that its existence is masked by bacterial diversity needed for breaking down different dietary carbohydrates. Furthermore, the objective of host control over the microbiota changes fundamentally as contents move from the small intestine into the colon, which will be discussed in sequence.
Small intestine: microbial population increase meets host control
Facultative anaerobic bacteria belonging to the families Lactobacillaceae (phylum Firmicutes) and Enterobacteriaceae (Proteobacteria) dominate the microbiota in the small intestine, with bacterial densities gradually increasing from 101–103 bacteria/ml in the duodenum to 104–107 bacteria/ml in the ileum.41 This bacterial community can de novo synthesize vitamin B-12 (cobalamin),42,43 which is thought to be beneficial because humans lack this biosynthetic capacity and thus rely on an exogenous source. Gastric intrinsic factor secreted by the gastric mucosa binds dietary or microbiota-derived cobalamin and promotes its absorption in the ileum.44
While the small-intestinal microbiota might aid in supplying the host with cobalamin, its community size needs to be controlled because the small intestine is the main site for absorption of simple sugars, amino acids, fatty acids, and glycerol, nutrients that are either present in our diet or generated by host enzymes through digestion of proteins, carbohydrates or fat. Since microbes can also consume these diet-derived nutrients, a large microbial community in the small intestine would compete with the host for food and produce malnutrition. Thus, to ensure the microbiota remains beneficial, the host has to prevent bacterial overgrowth in the small intestine. The host curbs bacterial numbers in the small intestine through gastric acid secretion, bowel motility, secretion of immunoglobulin and bacteriostatic properties of pancreatic and biliary secretions. Any illness that impairs these host control mechanisms puts a person at risk for small-intestinal bacterial overgrowth, which can result in malnutrition and weight loss.45
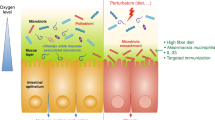
Another mechanism by which the host keeps control over the microbiota in the small intestine is the secretion of antimicrobial peptides by Paneth cells, specialized secretory epithelial cells located in the small-intestinal crypts. Paneth cells secrete an antimicrobial cocktail of defensins, lysozyme, phospholipase A2, and Reg3 lectins to contain gut microbes in close proximity to the mucosa (Fig. 2a).46 Their intensive secretory activity renders the extensive endoplasmic reticulum (ER) network of Paneth cells vulnerable to ER stress. Disruption of normal Paneth cell function by ER stress accompanies ileal Crohn’s disease, a form of inflammatory bowel disease.47 One risk factor for developing Crohn’s disease is childhood exposure to environmental factors present in geographical regions of high-disease incidence.48 Animal experiments suggest such environmental factors include dietary emulsifiers,49 a known trigger of ER stress in the intestinal epithelium.50,51 Genome-wide association studies identify genetic polymorphisms in the NOD252 and ATG16L1 genes53 as risk factors for developing Crohn’s disease. Interestingly, proteins encoded by the NOD2 and ATG16L1 genes are both functionally linked to ER stress,54,55 suggesting the corresponding risk alleles might jeopardize Paneth cell function. Consistent with ER stress-induced Paneth cell dysfunction, patients with Crohn’s disease exhibit decreased ileal synthesis of Paneth cell defensins,56 a trait linked to the NOD2 risk allele.57 In turn, disruption of normal Paneth cell function contributes to dysbiosis observed during Crohn’s disease (Fig. 2b).58
Paneth cell-mediated host control in the small intestine. a Antimicrobial peptides secreted by Paneth cells in the small-intestinal crypts check bacterial growth in close proximity to the epithelial surface, thereby maintaining homeostasis in the small intestine. b Genetic and environmental factors can co-operate to trigger ER stress, which results in intestinal inflammation and Paneth cell dysfunction. The latter reduces antimicrobial peptide secretion, thereby inducing dysbiosis characterized by an outgrowth of bacteria that reside in close proximity to the epithelial surface, which in turn can exacerbate intestinal inflammation
Direct evidence that Paneth cell-derived antimicrobial peptides shape the microbial community comes from experiments with transgenic mice expressing a human Paneth cell defensin, which results in the exclusion of segmented filamentous bacteria (phylum Firmicutes) from the microbial community.59 Segmented filamentous bacteria are intimately associated with the plasma membrane of murine epithelial cells, but are not a common constituent of the human small-intestinal microbiota,60 although it is not clear that synthesis of human Paneth cell defensin is responsible for their exclusion. However, Crohn’s disease is associated with a dysbiotic expansion of adherent-invasive Escherichia coli (family Enterobacteriaceae), which reside in close proximity to the mucosal surface.61 Thus, Paneth cell-derived antimicrobial peptides might serve to protect the epithelial surface, whereas impaired defensin synthesis accompanying Crohn’s disease might result in enhanced colonization of this niche (Fig. 2b). Adherent-invasive E. coli reduce expression of ATG16L1 in cultured epithelial cells, thereby impairing autophagy,62 an ER repair mechanism.63 This observation raises the possibility that dysbiosis enhances inflammation by exacerbating ER stress. The idea that dysbiosis exacerbates inflammation is supported by clinical data showing that antibiotic therapy can induce remission in active Crohn’s disease.64
Graft-versus-host disease (GvHD) is another illness associated with a disruption of Paneth cell function. GvHD is linked to reduced luminal defensin secretion and dysbiosis, which further accelerates the underlying illness.65,66 Interestingly, R-spondin 1-induced differentiation of intestinal stem cells into Paneth cells restores defensin secretion and prevents dysbiosis in a mouse model of GvHD.67 These data provide a proof of principle that rebalancing the gut microbiota by reinstating epithelial control over the microbial ecosystem represents a feasible therapeutic approach for restoring homeostasis.
Healthy guts exclude oxygen: how the host controls its colonic microbiota
As discussed above, host control in the small intestine is aimed at curbing bacterial numbers to avoid direct competition between host and microbiota for simple sugars and amino acids. In contrast, host control in the large intestine is no longer directed at limiting the size of the microbial community, as indicated by a steep rise in the bacterial density from 104–107 bacteria/ml in the ileum to 1011–1012 bacteria/ml in the colon.41 The large microbial community in the colon breaks down complex carbohydrates that cannot be digested by host enzymes in the upper gastrointestinal tract and reach the large bowel. In the absence of oxygen, obligate anaerobic bacteria catabolize complex carbohydrates into fermentation products, such as short-chain fatty acids, which are absorbed in the colon, thereby contributing to host nutrition.68,69 However, in the presence of oxygen, facultative anaerobic bacteria catabolize fermentation products to carbon dioxide,70 which is predicted to interfere with host nutrition. Thus, limiting oxygen availability in the colon helps the host to ensure the microbiota remains beneficial. To minimize the amount of oxygen emanating from the mucosal surface, the host maintains colonic epithelial cells (colonocytes) in a state of hypoxia (<1% oxygen).71 This host control mechanism ensures a dominance of obligate anaerobic bacteria in the colon without restricting diversity of the microbiota on the species level.72 The latter is functionally important, because bacterial diversity increases the probability of including species that can breakdown any given complex carbohydrate, thereby increasing the efficiency of obtaining short-chain fatty acids from a fiber-rich diet. Thus, while oxygen limitation in the colon controls the microbial ecosystem to ensure a dominance of obligate anaerobic bacteria,73 a lack of constraints on species diversity explains why this important host control mechanism is easily overlooked when a detailed analysis of fecal microbiota composition is performed.38
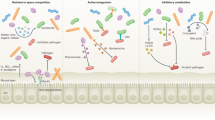
The host maintains anaerobiosis in the large bowel through a virtuous cycle that involves monitoring the presence of microbiota-derived fermentation products. Specifically, short-chain fatty acids produced by the microbial community of obligate anaerobic bacteria in the colon promote maturation and expansion of regulatory T cells74,75,76,77,78 and activate the epithelial butyrate-sensor PPAR-γ,73 which is highly expressed in differentiated colonocytes.79 Regulatory T cells and epithelial PPAR-γ-signaling act in concert to drive the energy metabolism of colonocytes toward oxygen consumption through β-oxidation, which renders the colonic surface hypoxic (Fig. 3a).73 In turn, epithelial hypoxia limits the amount of oxygen diffusing into the intestinal lumen, thereby maintaining a dominance of obligate anaerobic bacteria that breakdown fiber into short-chain fatty acids. This virtuous cycle provides stability73 and might contribute to microbiota resilience.80,81
Colonocyte-mediated host control in the large bowel. a Mature colonocytes obtain energy by PPAR-γ-activated mitochondrial β-oxidation of microbiota-derived butyrate (insert), a process that consumes oxygen (O2) and renders the epithelial surface hypoxic (<1% O2). Epithelial hypoxia drives anaerobiosis in the lumen, thereby ensuring a dominance of obligate anaerobic bacteria that convert fiber into short-chain fatty acids (SFAs), which serve host nutrition and suppress inflammation by promoting differentiation and expansion of regulatory T cells (Tregs). The color scale on the bottom left indicates O2 levels. b Inflammation and epithelial repair can shift the energy metabolism of colonocytes towards anaerobic glycolysis (insert). The consequent increase in epithelial oxygenation impairs the host’s ability to control the flow of oxygen into the intestinal lumen, which leads to dysbiosis characterized by a shift in the microbial community from obligate to facultative anaerobes
An impaired ability of the host to limit the flow of oxygen into the gut lumen is associated with a shift in the microbial community from obligate to facultative anaerobes,82 a common microbial signature of dysbiosis in the colon (Fig. 3b).83 For example, an expansion of facultative anaerobic Enterobacteriaceae is observed during ulcerative colitis, a form of inflammatory bowel disease affecting the colon.14 Based on this correlation, the “oxygen hypothesis” proposes that ulcerative colitis might be associated with a disruption in anaerobiosis.84 Recent work on mouse models of ulcerative colitis provides direct support for this idea. Metagenomic analysis of changes in the gut microbiota observed in dextran sulfate sodium (DSS)-induced colitis identifies oxygen respiration as a dominant microbial signature associated with inflammation.85 Citrobacter rodentium (family Enterobacteriaceae) triggers colonic crypt hyperplasia in mice, which mimics aspects of ulcerative colitis pathology.86 Colonic crypt hyperplasia is the result of excessive epithelial repair responses, leading to crypt elongation, the appearance of undifferentiated epithelial cells at the luminal surface and a consequent reduction in the numbers of goblet cells, which results in a thinning of the mucus layer. Colonic crypt hyperplasia shifts the colonocyte metabolism toward anaerobic glycolysis, thereby elevating epithelial oxygenation, which in turn drives a luminal expansion of C. rodentium through aerobic respiration.87 Similarly, aerobic respiration drives a luminal expansion of commensal E. coli during DSS-induced colitis.85 Thus, an expansion of Enterobacteriaceae during ulcerative colitis might be a microbial signature of a dysfunctional epithelium that no longer limits the flow of oxygen into the colonic lumen.88 The ensuing dysbiosis likely exacerbates intestinal inflammation, a conclusion supported by the clinical observation that ulcerative colitis can respond to antibiotic treatment.64
Above observations suggest that reestablishing anaerobiosis by restoring epithelial hypoxia could offer an alternative to FMTs for remediating dysbiosis. Unbeknownst to us, we might have already been harnessing this host control mechanism for therapeutic means for decades. Mesalazine, also known as 5-aminosalicylic acid (5-ASA), has been used for over 40 years as the first-line treatment for mild to moderate ulcerative colitis.89,90,91 5-ASA acts predominantly topical at the site of inflammation, especially within the colon,92,93,94 presumably by activating PPAR-γ-signaling in colonocytes.95 Since PPAR-γ-signaling is a strong driver of β-oxidation in colonocytes73 (Fig. 3a), 5-ASA likely aids in reestablishing epithelial hypoxia, thereby restoring host control mechanisms that limit oxygen availability in the colon.
When patients with ulcerative colitis develop severe exacerbation and require hospitalization, corticosteroids are the mainstay of therapy, while 5-ASA no longer has any effect on disease of this severity.96 This correlates with histopathological changes that include a loss of colonocytes (epithelial erosion),97 the therapeutic target of 5-ASA.95 Other histopathological indicators of severe acute ulcerative colitis include necrosis of tissue underlying areas of epithelial erosion (ulcers) and infiltration of neutrophils.97 Neutrophils undergoing a respiratory burst render the mucosal surface hypoxic.98 However, the respiratory burst generates alternative electron acceptors, such as nitrate and tetrathionate.99 Mouse models of colitis show that nitrate and tetrathionate can drive a colonic expansion of facultative anaerobic Enterobacteriaceae through anaerobic respiration.100,101,102,103,104 Thus, colitis is generally accompanied by an expansion of Enterobacteriaceae, but the mechanisms driving this expansion might differ between mild to moderate ulcerative colitis and severe acute ulcerative colitis.
The picture emerging from these studies is that during homeostasis the host keeps control over the colonic ecosystem by limiting the availability of exogenous respiratory electron acceptors to maintain anaerobiosis and check the growth of facultative anaerobic bacteria (Fig. 3a). Loss of this host control mechanism during colitis triggers a dysbiotic shift in the microbial community from obligate to facultative anaerobes (Fig. 3b).12 Epithelial PPAR-γ-signaling has emerged as a promising therapeutic target for restoring epithelial control over the colonic ecosystem to rebalance the microbiota.
Conclusions
The different digestive functions of the small and large intestines are each matched with a fitting host control mechanism that ensures the microbiota is compatible with and beneficial for host nutrition. An emerging theme throughout the gastrointestinal tract is the important role epithelial cells play in maintaining homeostasis by shaping the gut microbiota to be beneficial. The epithelial cell types exercising control over the microbiota differ between the small and large bowel. While Paneth cells secrete antimicrobial peptides to control the microbiota in the small intestine (Fig. 2),58 colonocytes limit the flow of oxygen into the gut lumen to shape the microbiota composition in the large intestine (Fig. 3).88 Since the human host already has ways to regulate the microbiota, reinstating epithelial control over the gut ecosystem offers an alternative to targeting the microbes themselves for remediating dysbiosis. Harnessing epithelial control for therapeutic means therefore represents a promising novel strategy for rebalancing the microbiota in a broad spectrum of human diseases.
References
Koch, R. Die Aetiologie derTuberkulose. Berl. Klin. Wochenschr. 15, 221–230 (1882).
Round, J. L. & Mazmanian, S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 (2009).
Packey, C. D. & Sartor, R. B. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr. Opin. Infect. Dis. 22, 292–301 (2009).
Neville, B. A., Forster, S. C. & Lawley, T. D. Commensal Koch’s postulates: establishing causation in human microbiota research. Curr. Opin. Microbiol. 42, 47–52 (2017).
Halkjaer, S. I., Boolsen, A. W., Gunther, S., Christensen, A. H. & Petersen, A. M. Can fecal microbiota transplantation cure irritable bowel syndrome? World J. Gastroenterol. 23, 4112–4120 (2017).
Choi, H. H. & Cho, Y. S. Fecal microbiota transplantation: current applications, effectiveness, and future perspectives. Clin. Endosc. 49, 257–265 (2016).
Kelly, C. R. et al. Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am. J. Gastroenterol. 109, 1065–1071 (2014).
Frank, D. N., Zhu, W., Sartor, R. B. & Li, E. Investigating the biological and clinical significance of human dysbioses. Trends Microbiol. 19, 427–434 (2011).
Aroniadis, O. C. & Brandt, L. J. Fecal microbiota transplantation: past, present and future. Curr. Opin. Gastroenterol. 29, 79–84 (2013).
Sbahi, H. & Di Palma, J. A. Faecal microbiota transplantation: applications and limitations in treating gastrointestinal disorders. BMJ Open Gastroenterol. 3, e000087 (2016).
Konig, J. et al. Consensus report: faecal microbiota transfer - clinical applications and procedures. Aliment. Pharmacol. Ther. 45, 222–239 (2017).
Byndloss, M. X. & Bäumler, A. J. The germ-oran theory of non-communicable diseases. Nat. Rev. Microbiol., In press (2017).
Foster, K. R., Schluter, J., Coyte, K. Z. & Rakoff-Nahoum, S. The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51 (2017).
Morgan, X. C. et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79 (2012).
Yoshimoto, S. et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013).
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011).
Hsiao, E. Y. et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013).
Minter, M. R. et al. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer’s disease. Sci. Rep. 6, 30028 (2016).
Ruby, E. G. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50, 591–624 (1996).
Wei, S. L. & TYoung, R. E. Development of symbiotic bacterial bioluminescence in a nearshore cephalopod, Euprymna scolopes. Mar. Biol. 103, 541–546 (1989).
Venter, J. C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004).
Chen, F. et al. Bactericidal permeability-increasing proteins shape host-microbe interactions. MBio 8, (2017). https://doi.org/10.1128/mBio.00040-17
Heath-Heckman, E. A. et al. Shaping the microenvironment: evidence for the influence of a host galaxin on symbiont acquisition and maintenance in the squid-Vibrio symbiosis. Environ. Microbiol. 16, 3669–3682 (2014).
Troll, J. V. et al. Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ. Microbiol. 12, 2190–2203 (2010).
Studer, S. V. et al. Non-native acylated homoserine lactones reveal that LuxIR quorum sensing promotes symbiont stability. Environ. Microbiol. 16, 2623–2634 (2014).
Kremer, N. et al. The dual nature of haemocyanin in the establishment and persistence of the squid-vibrio symbiosis. Proc. Biol. Sci. 281, 20140504 (2014).
Ravel, J. et al. Vaginal microbiome of reproductive-age women. Proc. Natl Acad. Sci. USA 108, 4680–4687 (2011).
Chen, C. et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 8, 875 (2017).
Muhleisen, A. L. & Herbst-Kralovetz, M. M. Menopause and the vaginal microbiome. Maturitas 91, 42–50 (2016).
Spear, G. T. et al. Human alpha-amylase present in lower-genital-tract mucosal fluid processes glycogen to support vaginal colonization by Lactobacillus. J. Infect. Dis. 210, 1019–1028 (2014).
Boris, S. & Barbes, C. Role played by lactobacilli in controlling the population of vaginal pathogens. Microbes Infect. 2, 543–546 (2000).
Barile, D. & Rastall, R. A. Human milk and related oligosaccharides as prebiotics. Curr. Opin. Biotechnol. 24, 214–219 (2013).
Roger, L. C., Costabile, A., Holland, D. T., Hoyles, L. & McCartney, A. L. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156, 3329–3341 (2010).
Sela, D. A. et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl Acad. Sci. USA 105, 18964–18969 (2008).
Sela, D. A. & Mills, D. A. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 18, 298–307 (2010).
Watkins, C., Stanton, C., Ryan, C. A. & Ross, R. P. Microbial therapeutics designed for infant health. Front. Nutr. 4, (2017). https://doi.org/10.3389/fnut.2017.00048
Tap, J. et al. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11, 2574–2584 (2009).
Turnbaugh, P. J. et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14 (2009).
Thursby, E. & Juge, N. Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836 (2017).
Sommer, F. & Backhed, F. Know your neighbor: microbiota and host epithelial cells interact locally to control intestinal function and physiology. Bioessays 38, 455–464 (2016).
Taranto, M. P., Vera, J. L., Hugenholtz, J., De Valdez, G. F. & Sesma, F. Lactobacillus reuteri CRL1098 produces cobalamin. J. Bacteriol. 185, 5643–5647 (2003).
Lawrence, J. G. & Roth, J. R. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics 142, 11–24 (1996).
Alpers, D. H. & Russell-Jones, G. Gastric intrinsic factor: the gastric and small intestinal stages of cobalamin absorption. a personal journey. Biochimie 95, 989–994 (2013).
Bures, J. et al. Small intestinal bacterial overgrowth syndrome. World J. Gastroenterol. 16, 2978–2990 (2010).
Bevins, C. L. & Salzman, N. H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368 (2011).
Kaser, A. & Blumberg, R. S. ATG16L1 Crohn’s disease risk stresses the endoplasmic reticulum of Paneth cells. Gut 63, 1038–1039 (2014).
Gearry, R. B. IBD and environment: are there differences between east and west. Dig. Dis. 34, 84–89 (2016).
Chassaing, B. et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96 (2015).
Gulhane, M. et al. High fat diets induce colonic epithelial cell stress and inflammation that is reversed by IL-22. Sci. Rep. 6, 28990 (2016).
Fu, S. et al. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature 473, 528–531 (2011).
Hampe, J. et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet 357, 1925–1928 (2001).
Hampe, J. et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211 (2007).
Keestra-Gounder, A. M. et al. NOD1 and NOD2 signalling links ER stress with inflammation. Nature 532, 394–397 (2016).
Deuring, J. J. et al. Genomic ATG16L1 risk allele-restricted Paneth cell ER stress in quiescent Crohn’s disease. Gut 63, 1081–1091 (2014).
Wehkamp, J. et al. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal alpha-defensin expression. Gut 53, 1658–1664 (2004).
Bevins, C. L., Stange, E. F. & Wehkamp, J. Decreased Paneth cell defensin expression in ileal Crohn’s disease is independent of inflammation, but linked to the NOD2 1007fs genotype. Gut 58, 882–883 (2009). discussion 883–884.
Salzman, N. H. & Bevins, C. L. Dysbiosis--a consequence of Paneth cell dysfunction. Semin. Immunol. 25, 334–341 (2013).
Salzman, N. H. et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11, 76–83 (2010).
Sczesnak, A. et al. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe 10, 260–272 (2011).
Darfeuille-Michaud, A. et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127, 412–421 (2004).
Nguyen, H. T. et al. Crohn’s disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology 146, 508–519 (2014).
Tschurtschenthaler, M. et al. Defective ATG16L1-mediated removal of IRE1alpha drives Crohn’s disease-like ileitis. J. Exp. Med. 214, 401–422 (2017).
Khan, K. J. et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am. J. Gastroenterol. 106, 661–673 (2011).
Jenq, R. R. et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J. Exp. Med. 209, 903–911 (2012).
Eriguchi, Y. et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of alpha-defensins. Blood 120, 223–231 (2012).
Hayase, E. et al. R-Spondin1 expands Paneth cells and prevents dysbiosis induced by graft-versus-host disease. J. Exp. Med., Online early, (2017). https://doi.org/10.1084/jem.20170418
den Besten, G. et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340 (2013).
Velazquez, O. C., Lederer, H. M. & Rombeau, J. L. Butyrate and the colonocyte. Production, absorption, metabolism, and therapeutic implications. Adv. Exp. Med. Biol. 427, 123–134 (1997).
Spiga, L. et al. An oxidative central metabolism enables salmonella to utilize microbiota-derived succinate. Cell. Host. Microbe 22, 291–301 e296 (2017).
Zheng, L., Kelly, C. J. & Colgan, S. P. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. Am. J. Physiol. Cell Physiol. 309, C350–360 (2015).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Byndloss, M. X. et al. Microbiota-activated PPAR-g signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575 (2017).
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Smith, P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Singh, N. et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139 (2014).
Lefebvre, M. et al. Peroxisome proliferator-activated receptor gamma is induced during differentiation of colon epithelium cells. J. Endocrinol. 162, 331–340 (1999).
Relman, D. A. The human microbiome: ecosystem resilience and health. Nutr. Rev. 70, S2–9 (2012).
Sommer, F., Anderson, J. M., Bharti, R., Raes, J. & Rosenstiel, P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 15, 630–638 (2017).
Rivera-Chavez, F., Lopez, C. A. & Baumler, A. J. Oxygen as a driver of gut dysbiosis. Free Radic. Biol. Med. 105, 93–101 (2017).
Shin, N. R., Whon, T. W. & Bae, J. W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503 (2015).
Rigottier-Gois, L. Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J. 7, 1256–1261 (2013).
Hughes, E. R. et al. Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host. Microbe 21, 208–219 (2017).
Luperchio, S. A. & Schauer, D. B. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect. 3, 333–340 (2001).
Lopez, C. A. et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353, 1249–1253 (2016).
Litvak, Y., Byndloss, M. X., Tsolis, R. M. & Baumler, A. J. Dysbiotic proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr. Opin. Microbiol. 39, 1–6 (2017).
Azad Khan, A. K., Piris, J. & Truelove, S. C. An experiment to determine the active therapeutic moiety of sulphasalazine. Lancet 2, 892–895 (1977).
Iacucci, M., de Silva, S. & Ghosh, S. Mesalazine in inflammatory bowel disease: a trendy topic once again? Can. J. Gastroenterol. 24, 127–133 (2010).
Criscuoli, V., Modesto, I., Orlando, A. & Cottone, M. Mesalazine for the treatment of inflammatory bowel disease. Expert. Opin. Pharmacother. 14, 1669–1678 (2013).
Brogden, R. N. & Sorkin, E. M. Mesalazine. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in chronic inflammatory bowel disease. Drugs 38, 500–523 (1989).
Greenfield, S. M., Punchard, N. A., Teare, J. P. & Thompson, R. P. Review article: the mode of action of the aminosalicylates in inflammatory bowel disease. Aliment. Pharmacol. Ther. 7, 369–383 (1993).
Zhou, S. Y. et al. Intestinal metabolism and transport of 5-aminosalicylate. Drug Metab. Dispos. 27, 479–485 (1999).
Rousseaux, C. et al. Intestinal antiinflammatory effect of 5-aminosalicylic acid is dependent on peroxisome proliferator-activated receptor-gamma. J. Exp. Med. 201, 1205–1215 (2005).
Kedia, S., Ahuja, V. & Tandon, R. Management of acute severe ulcerative colitis. World J. Gastrointest. Pathophysiol. 5, 579–588 (2014).
Langner, C. et al. Histology of microscopic colitis-review with a practical approach for pathologists. Histopathology 66, 613–626 (2015).
Campbell, E. L. et al. Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity 40, 66–77 (2014).
Winter, S. E., Lopez, C. A. & Baumler, A. J. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 14, 319–327 (2013).
Winter, S. E. et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010).
Lopez, C. A. et al. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. MBio 3, e00143–12 (2012).
Winter, S. E. et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013).
Lopez, C. A., Rivera-Chavez, F., Byndloss, M. X. & Baumler, A. J. The periplasmic nitrate reductase NapABC supports luminal growth of salmonella enterica serovar typhimurium during colitis. Infect. Immun. 83, 3470–3478 (2015).
Kamdar, K. et al. Genetic and metabolic signals during acute enteric bacterial infection alter the microbiota and drive progression to chronic inflammatory disease. Cell Host Microbe 19, 21–31 (2016).
Authors contributions
M.X.B. and A.J.B. wrote the manuscript. S.R.P. generated scientific Illustrations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Byndloss, M.X., Pernitzsch, S.R. & Bäumler, A.J. Healthy hosts rule within: ecological forces shaping the gut microbiota. Mucosal Immunol 11, 1299–1305 (2018). https://doi.org/10.1038/s41385-018-0010-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-018-0010-y