Abstract
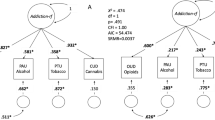
Substance use disorders (SUDs) incur serious social and personal costs. The risk for SUDs is complex, with risk factors ranging from social conditions to individual genetic variation. We examined whether models that include a clinical/environmental risk index (CERI) and polygenic scores (PGS) are able to identify individuals at increased risk of SUD in young adulthood across four longitudinal cohorts for a combined sample of N = 15,134. Our analyses included participants of European (NEUR = 12,659) and African (NAFR = 2475) ancestries. SUD outcomes included: (1) alcohol dependence, (2) nicotine dependence; (3) drug dependence, and (4) any substance dependence. In the models containing the PGS and CERI, the CERI was associated with all three outcomes (ORs = 01.37–1.67). PGS for problematic alcohol use, externalizing, and smoking quantity were associated with alcohol dependence, drug dependence, and nicotine dependence, respectively (OR = 1.11–1.33). PGS for problematic alcohol use and externalizing were also associated with any substance dependence (ORs = 1.09–1.18). The full model explained 6–13% of the variance in SUDs. Those in the top 10% of CERI and PGS had relative risk ratios of 3.86–8.04 for each SUD relative to the bottom 90%. Overall, the combined measures of clinical, environmental, and genetic risk demonstrated modest ability to distinguish between affected and unaffected individuals in young adulthood. PGS were significant but added little in addition to the clinical/environmental risk index. Results from our analysis demonstrate there is still considerable work to be done before tools such as these are ready for clinical applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All data sources are described in the manuscript and supplemental information. No new data were collected. Only data from existing studies or study cohorts were analyzed, some of which have restricted access to protect the privacy of the study participants. Add Health genetic data obtained through dbGaP (Study Accession: phs001367.v1.p1). Instructions on gaining access to Add Health restricted use data can be found at: https://data.cpc.unc.edu/projects/2/view. COGA genetic data available through dbGaP (Study Accession: phs000763.v1.p1). Instructions for access to ALSPAC data available at: http://www.bristol.ac.uk/alspac/researchers/access/. The process for obtaining the GWAS summary statistics used in these analyses are described in the corresponding original GWAS publications.
Code availability
No custom algorithms or software was developed in this study. All code is available by request from the corresponding author. Polygenic scores generated using PRS-CSx (https://github.com/getian107/PRScsx). All primary analyses completed in R 4.1.0 using the data.table(1.14.0), pROC (1.18.0), lme4 (1.1–27.1), DescTools (0.99.45), sandwich (3.0–2), and base packages.
References
U.S. Overdose deaths in 2021 increased half as much as in 2020 - but are still up 15%. https://www.cdc.gov/nchs/pressroom/nchs_press_releases/2022/202205.htm. Accessed 15 May 2022.
Degenhardt L, Charlson F, Ferrari A, Santomauro D, Erskine H, Mantilla-Herrara A, et al. The global burden of disease attributable to alcohol and drug use in 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Psychiatry 2018;5:987–1012.
Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, Brewer RD. National and state costs of excessive alcohol consumption. Am J Prev Med. 2010;2015:e73–e79.
National Drug Intelligence Center. National drug threat assessment. 2019. Washington, DC: United States Department of Justice; 2011.
Verhulst B, Neale MC, Kendler KS. The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med. 2015;45:1061–72.
Verweij KJH, Zietsch BP, Lynskey MT, Medland SE, Neale MC, Martin NG, et al. Genetic and environmental influences on cannabis use initiation and problematic use: a meta-analysis of twin studies. Addiction. 2010;105:417–30.
Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry 2003;160:687–95.
Galea S, Nandi A, Vlahov D. The social epidemiology of substance use. Epidemiol Rev. 2004;26:36–52.
Barr PB. Neighborhood conditions and trajectories of alcohol use and misuse across the early life course. Health Place. 2018;51:36–44.
Barr PB, Silberg J, Dick DM, Maes HH. Childhood socioeconomic status and longitudinal patterns of alcohol problems: variation across etiological pathways in genetic risk. Soc Sci Med. 2018;209:51–58.
Meier MH, Hall W, Caspi A, Belsky DW, Cerda M, Harrington HL, et al. Which adolescents develop persistent substance dependence in adulthood? Using population-representative longitudinal data to inform universal risk assessment. Psychol Med. 2016;46:877–89.
Schaefer JD, Jang SK, Clark DA, Deak JD, Hicks BM, Iacono WG, et al. Associations between polygenic risk of substance use and use disorder and alcohol, cannabis, and nicotine use in adolescence and young adulthood in a longitudinal twin study. Psychol Med. 2021:1–11. (Online available ahead of printing).
Deak JD, Clark DA, Liu M, Schaefer JD, Jang SK, Durbin CE, et al. Alcohol and nicotine polygenic scores are associated with the development of alcohol and nicotine use problems from adolescence to young adulthood. Addiction. 2022;117:1117–27.
Barr PB, Ksinan A, Su J, Johnson EC, Meyers JL, Wetherill L, et al. Using polygenic scores for identifying individuals at increased risk of substance use disorders in clinical and population samples. Transl Psychiatry 2020;10:196.
Kinreich S, Meyers JL, Maron-Katz A, Kamarajan C, Pandey AK, Chorlian DB, et al. Predicting risk for Alcohol Use Disorder using longitudinal data with multimodal biomarkers and family history: a machine learning study. Mol Psychiatry 2021;26:1133–41.
Gu F, Chen TH, Pfeiffer RM, Fargnoli MC, Calista D, Ghiorzo P, et al. Combining common genetic variants and non-genetic risk factors to predict risk of cutaneous melanoma. Hum Mol Genet. 2018. https://doi.org/10.1093/hmg/ddy282.
O’Sullivan JW, Shcherbina A, Justesen JM, Turakhia M, Perez M, Wand H, et al. Combining clinical and polygenic risk improves stroke prediction among individuals with atrial fibrillation. Circ Genom Precis Med. 2021;14:339–47.
Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE, et al. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry 2005;62:593.
Harris KM, Halpern CT, Haberstick BC, Smolen A. The National Longitudinal Study of Adolescent Health (Add Health) sibling pairs data. Twin Res Hum Genet. 2013;16:391–8.
Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, et al. Cohort profile: The ‘Children of the 90s’-The index offspring of the avon longitudinal study of parents and children. Int J Epidemiol. 2013. https://doi.org/10.1093/ije/dys064.
Fraser A, Macdonald-wallis C, Tilling K, Boyd A, Golding J, Davey smith G, et al. Cohort profile: the avon longitudinal study of parents and children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42:97–110.
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)-A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inf. 2009;42:377–81.
Northstone K, Lewcock M, Groom A, Boyd A, Macleod J, Timpson N, et al. The Avon Longitudinal Study of Parents and Children (ALSPAC): an update on the enrolled sample of index children in 2019 [version 1; peer review: 2 approved]. Wellcome Open Res. 2019;4:51.
Edenberg HJ. The collaborative study on the genetics of alcoholism: an update. Alcohol Res Health. 2002;26:214–8.
Begleiter H. The collaborative study on the genetics of alcoholism. Alcohol Health Res World. 1995;19:228.
Bucholz KK, McCutcheon VV, Agrawal A, Dick DM, Hesselbrock VM, Kramer JR, et al. Comparison of parent, peer, psychiatric, and cannabis use influences across stages of offspring alcohol involvement: evidence from the COGA prospective study. Alcohol Clin Exp Res. 2017;41:359–68.
Rose RJ, Salvatore JE, Aaltonen S, Barr PB, Bogl LH, Byers HA et al. FinnTwin12 Cohort: An Updated Review. Twin Res Hum Genet. 2019;22:302–11.
Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–27.
Hughes K, Bellis MA, Hardcastle KA, Sethi D, Butchart A, Mikton C, et al. The effect of multiple adverse childhood experiences on health: a systematic review and meta-analysis. Lancet Public Health. 2017. https://doi.org/10.1016/S2468-2667(17)30118-4.
Sher KJ, Grekin ER, Williams NA. The development of alcohol use disorders. Annu Rev Clin Psychol. 2005;1:493–523.
Karlsson Linner R, Mallard TT, Barr PB, Sanchez-Roige S, Madole JW, Driver MN, et al. Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat Neurosci. 2021;24:1367–76.
Levey DF, Stein MB, Wendt FR, Pathak GA, Zhou H, Aslan M, et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat Neurosci. 2021. https://doi.org/10.1038/s41593-021-00860-2.
Zhou H, Sealock JM, Sanchez-Roige S, Clarke TK, Levey DF, Cheng Z, et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci. 2020. https://doi.org/10.1038/s41593-020-0643-5.
Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–44.
Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun. 2019;10:1499.
Quach BC, Bray MJ, Gaddis NC, Liu M, Palviainen T, Minica CC, et al. Expanding the genetic architecture of nicotine dependence and its shared genetics with multiple traits. Nat Commun. 2020;11:5562.
Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;2022:1–13.
Bigdeli TB, Fanous AH, Li Y, Rajeevan N, Sayward F, Genovese G, et al. Genome-wide association studies of schizophrenia and bipolar disorder in a diverse cohort of US Veterans. Schizophr Bull. 2020. https://doi.org/10.1093/schbul/sbaa133.
Barr PB, Dick DM. The genetics of externalizing problems. Curr Top Behav Neurosci. 2020;47:93–112.
Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WGWG, McGue M. Etiological connections among substance dependence, antisocial behavior and personality: modeling the externalizing spectrum. J Abnorm Psychol. 2002;111:411–24.
Kendler KS, Myers J. The boundaries of the internalizing and externalizing genetic spectra in men and women. Psychol Med. 2014;44:647–55.
Polimanti R, Peterson RE, Ong JS, MacGregor S, Edwards AC, Clarke TK, et al. Evidence of causal effect of major depression on alcohol dependence: findings from the psychiatric genomics consortium. Psychol Med. 2019. https://doi.org/10.1017/S0033291719000667.
Johnson EC, Demontis D, Thorgeirsson TE, Walters RK, Polimanti R, Hatoum AS, et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry 2020. https://doi.org/10.1016/S2215-0366(20)30339-4.
Zhou H, Rentsch CT, Cheng Z, Kember RL, Nunez YZ, Sherva RM, et al. Association of OPRM1 Functional Coding Variant With Opioid Use Disorder: A Genome-Wide Association Study. JAMA Psychiatry 2020. https://doi.org/10.1001/jamapsychiatry.2020.1206.
Kendler KS, Gardner C, Dick DM. Predicting alcohol consumption in adolescence from alcohol- specific and general externalizing genetic risk factors, key environmental exposures and their interaction. Psychol Med. 2011;41:1507–16.
Meyers JL, Salvatore JE, Vuoksimaa E, Korhonen T, Pulkkinen L, Rose RJ, et al. Genetic influences on alcohol use behaviors have diverging developmental trajectories: a prospective study among male and female twins. Alcohol Clin Exp Res. 2014;38:2869–77.
Barr PB, Mallard TT, Sanchez-Roige S, Poore HE, Linnér RK, Collaborators C, et al. Parsing genetically influenced risk pathways: genetic loci impact problematic alcohol use via externalizing and specific risk. 2021. https://www.medrxiv.org/content/10.1101/2021.07.20.21260861v1.
Sanchez-Roige S, Palmer AA, Clarke TK. Recent efforts to dissect the genetic basis of alcohol use and abuse. Biol Psychiatry 2020;87:609–18.
Walters RK, Polimanti R, Johnson EOECEO, McClintick JN, Adams MJ, Adkins AE, et al. Trans-ancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 2018;21:1656–69.
Dick DM, Barr P, Guy M, Nasim A, Scott D. Review: genetic research on alcohol use outcomes in African American populations: A review of the literature, associated challenges, and implications. Am J Addictions. 2017;26:486–93.
Mills MC, Rahal C. A scientometric review of genome-wide association studies. Commun Biol. 2019;2:9.
Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100:635–49.
Duncan L, Shen H, Gelaye B, Meijsen J, Ressler K, Feldman M, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019. https://doi.org/10.1038/s41467-019-11112-0.
Ruan Y, Lin Y-F, Feng Y-CA, Chen C-Y, Lam M, Guo Z, et al. Improving polygenic prediction in ancestrally diverse populations. Nat Genet. 2022. https://doi.org/10.1038/s41588-022-01054-7.
Curran PJ, Hussong AM. Integrative data analysis: the simultaneous analysis of multiple data sets. Psychol Methods. 2009;14:81–100.
Cameron CA, Gelbach JB, Miller DL. Robust inference with multiway clustering. J Bus Economic Stat. 2011;29:238–49.
Nagelkerke NJD. A note on a general definition of the coefficient of determination. Biometrika. 1991;78:691–2.
Hasin DS, Grant BF. The National Epidemiologic Survey on Alcohol and Related Conditions (NESARC) Waves 1 and 2: review and summary of findings. Soc Psychiatry Psychiatr Epidemiol. 2015;50:1609–40.
Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young AI, Thorgeirsson TE, et al. The nature of nurture: Effects of parental genotypes. Science. 2018;359:424–8.
Conrod PJ, O’Leary-Barrett M, Newton N, Topper L, Castellanos-Ryan N, MacKie C, et al. Effectiveness of a selective, personality-targeted prevention program for adolescent alcohol use and misuse: a cluster randomized controlled trial. JAMA Psychiatry. 2013;70:334–42.
Garg A, Boynton-Jarrett R, Dworkin PH. Avoiding the unintended consequences of screening for social determinants of health. JAMA. 2016;316:813–4.
Davidson KW, McGinn T. Screening for social determinants of health: the known and unknown. JAMA. 2019;322:1037–8.
Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–91.
Williams DR, Mohammed SA, Leavell J, Collins C. Race, socioeconomic status, and health: complexities, ongoing challenges, and research opportunities. Ann N Y Acad Sci. 2010;1186:69–101.
Acknowledgements
Research reported in this publication was supported by the National Institute on Alcohol Abuse and Alcoholism and the National Institute of Drug Abuse of the National Institutes of Health under award numbers R01AA015416, R01DA050721, R01DA042090, and K02AA018755; the Academy of Finland (grants 100499, 205585, 118555, 141054, 265240, 308248, 308698 and 312073); and the Scientific and Technological Research Council of Turkey (TÜBİTAK) under award number 114C117 (FA); and the Sigrid Juselius Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding bodies. This research also used summary data from the Psychiatric Genomics Consortium (PGC), the Million Veterans Program (MVP), the GWAS and Sequencing Consortium for Alcohol and Nicotine (GSCAN), UK Biobank, the Genomic Psychiatry Cohort (GPC) and 23andMe, Inc. We would like to thank the many studies that made these consortia possible, the researchers involved, and the participants in those studies, without whom this effort would not be possible. We would also like to thank the research participants and employees of 23andMe. The Externalizing Consortium: Principal Investigators: Danielle M. Dick, Philipp Koellinger, K. Paige Harden, Abraham A. Palmer. Lead Analysts: Richard Karlsson Linnér, Travis T. Mallard, Peter B. Barr, Sandra Sanchez-Roige. Significant Contributors: Irwin D. Waldman. The Externalizing Consortium has been supported by the National Institute on Alcohol Abuse and Alcoholism (R01AA015416 - administrative supplement), and the National Institute on Drug Abuse (R01DA050721). Additional funding for investigator effort has been provided by K02AA018755, U10AA008401, P50AA022537, as well as a European Research Council Consolidator Grant (647648 EdGe to Koellinger). The content is solely the responsibility of the authors and does not necessarily represent the official views of the above funding bodies. Add Health: Add Health is directed by Robert A. Hummer and funded by the National Institute on Aging cooperative agreements U01 AG071448 (Hummer) and U01AG071450 (Aiello and Hummer) at the University of North Carolina at Chapel Hill. Waves I–V data are from the Add Health Program Project, grant P01 HD31921 (Harris) from Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), with cooperative funding from 23 other federal agencies and foundations. Add Health was designed by J. Richard Udry, Peter S. Bearman, and Kathleen Mullan Harris at the University of North Carolina at Chapel Hill. ALSPAC: We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. The UK Medical Research Council and Wellcome (Grant ref: 217065/Z/19/Z) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors, and Peter Barr and Danielle Dick will serve as guarantors for the contents of this paper. A comprehensive list of grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf); This research was specifically funded by the Medical Research Council (MRC) under grants MR/L022206/1, MR/M006727/1, and G0800612/86812; the Wellcome Trust under grant 086684; and the National Institute on Alcohol Abuse and Alcoholism under 5R01AA018333–05. GWAS data was generated by Sample Logistics and Genotyping Facilities at Wellcome Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe. COGA: We thank The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, T. Foroud; Scientific Director, A. Agrawal; Translational Director, D. Dick, includes eleven different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, T. Foroud, Y. Liu, M.H. Plawecki); University of Iowa Carver College of Medicine (S. Kuperman, J. Kramer); SUNY Downstate Health Sciences University (B. Porjesz, J. Meyers, C. Kamarajan, A. Pandey); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz, A. Agrawal); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield, R. Hart, J. Salvatore); The Children’s Hospital of Philadelphia, University of Pennsylvania (L. Almasy); Virginia Commonwealth University (D. Dick); Icahn School of Medicine at Mount Sinai (A. Goate, P. Slesinger); and Howard University (D. Scott). Other COGA collaborators include: L. Bauer (University of Connecticut); J. Nurnberger Jr., L. Wetherill, X., Xuei, D. Lai, S. O’Connor, (Indiana University); G. Chan (University of Iowa; University of Connecticut); D.B. Chorlian, J. Zhang, P. Barr, S. Kinreich, G. Pandey (SUNY Downstate); N. Mullins (Icahn School of Medicine at Mount Sinai); A. Anokhin, S. Hartz, E. Johnson, V. McCutcheon, S. Saccone (Washington University); J. Moore, Z. Pang, S. Kuo (Rutgers University); A. Merikangas (The Children’s Hospital of Philadelphia and University of Pennsylvania); F. Aliev (Virginia Commonwealth University); H. Chin and A. Parsian are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting- Kai Li, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). All code necessary to replicate this study is available upon request. We are grateful Drs. Amy Taylor and Joëlle A. Pasman for providing GWAS summary statistics with ALSPAC samples removed.
Author information
Authors and Affiliations
Contributions
PBB, SIK, MND, and DMD conceived the study. DMD oversaw the study. PBB led the writing of the manuscript, with substantive contributions to the writing from DMD, SIK, and MND. PBB was the lead analyst and prepared data in Add Health and FinnTwin12. SIK prepared data in COGA. MND prepared data in ALSPAC. RKL, FA, and JM provided GWAS summary statistics. MS, KPH, BP, KB, JK, AL, HJE, MHP, and AAP provided helpful advice and feedback on various aspects of the study design. All authors contributed to and critically reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Barr, P.B., Driver, M.N., Kuo, S.IC. et al. Clinical, environmental, and genetic risk factors for substance use disorders: characterizing combined effects across multiple cohorts. Mol Psychiatry 27, 4633–4641 (2022). https://doi.org/10.1038/s41380-022-01801-6
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-022-01801-6